Fast and Accurate Peptide - MHC Structure Prediction via an Equivariant Diffusion Model
Fast and Accurate Peptide - MHC Structure Prediction via an Equivariant Diffusion Model
Fruehbuss, D.; Baakman, C.; Teusink, S.; Bekkers, E.; Jegelka, S.; Xue, L. C.
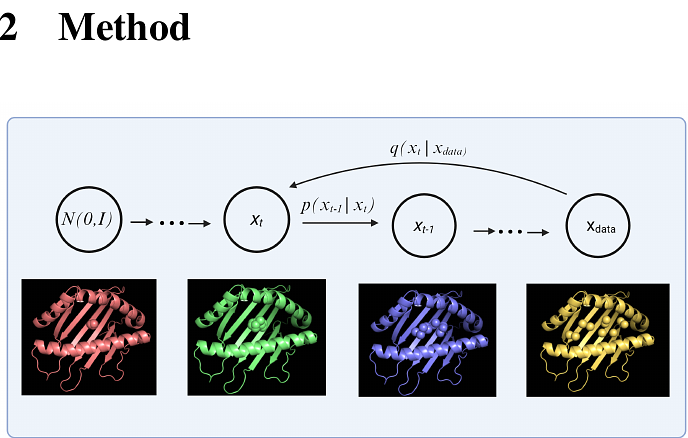
AbstractAccurate modeling of peptide-MHC (Major Histocompatibility Complex) structures is critical for the development of personalized cancer vaccines and T-cell therapies, as MHC proteins present peptides on the cell surface for immune recognition. Here, we introduce MHC-Diff, a specialized SE(3)-equivariant diffusion model leveraging Geometric Deep Learning to predict the 3D C-alpha atom structures of peptide-MHC complexes with high accuracy. Unlike previous deep learning models that generate a single static structure, our probabilistic approach samples multiple diverse candidates, capturing the inherent flexibility of peptide-MHC binding. Validated on the Pandora benchmark and experimental X-ray crystallography data, MHC-Diff achieves sub-angstrom accuracy, outperforming existing methods by a large margin while matching the inference speed of the fastest available techniques. By enabling rapid and highly accurate structure prediction across diverse peptide lengths and MHC alleles, MHC-Diff provides a powerful new tool for accelerating the design of next-generation cancer vaccines and T-cell therapies.