Robust Prediction of Multiple Protein Conformations with Entropy Guidance
Robust Prediction of Multiple Protein Conformations with Entropy Guidance
Wu, D.; Feng, L.
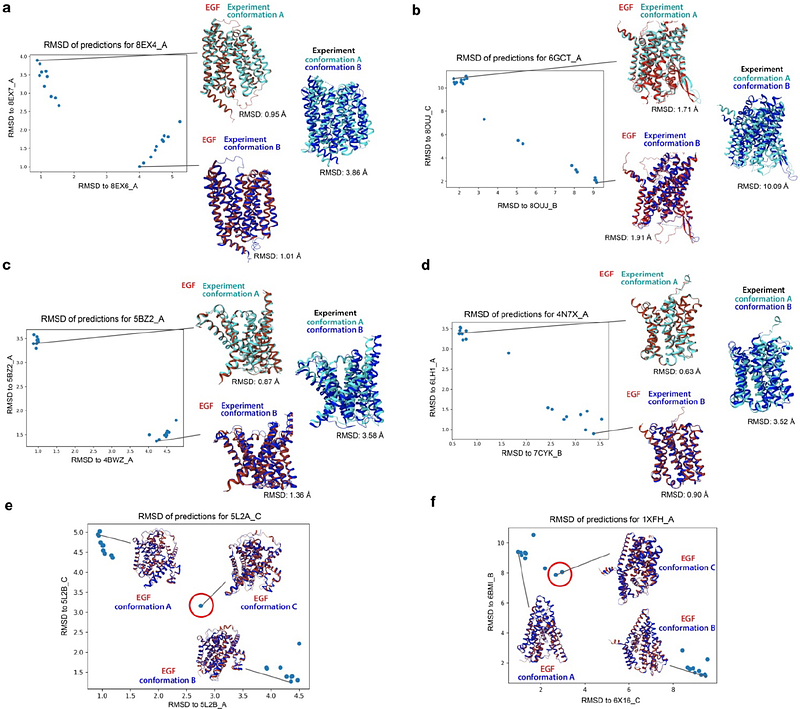
AbstractDeep learning approaches, exemplified by AlphaFold, have made enormous progress in addressing the grand challenge of protein structure prediction. However, these methods typically predict only a single conformation, while accurately predicting multiple conformations, which are essential for the functions and regulations of many protein classes, remains challenging. Here, we demonstrate that AlphaFold2 has internal knowledge of alternative protein conformations, which can be effectively extracted by learning modifications to its intermediate hidden states using a negative entropy loss. Our resulting method, Entropy Guided Fold (EGF), achieves a success rate of over 80% in accurately predicting two conformations of 37 tested membrane proteins within a 2.0 angstrom RMSD threshold, with similar performance for proteins both within and outside AlphaFold2's training dataset. In addition, our method successfully reveals three or more conformational states in several cases. We have also developed a post-processing strategy to effectively select accurate predictions. Overall, our method provides an efficient and robust approach for predicting multiple conformations of target proteins, with broad potential applications in mechanistic studies and translational applications.