DrugDomain2.0: comprehensive database of protein domains-ligands/drugs interactions across the whole Protein Data Bank
DrugDomain2.0: comprehensive database of protein domains-ligands/drugs interactions across the whole Protein Data Bank
Medvedev, K. E.; Schaeffer, R. D.; Grishin, N. V.
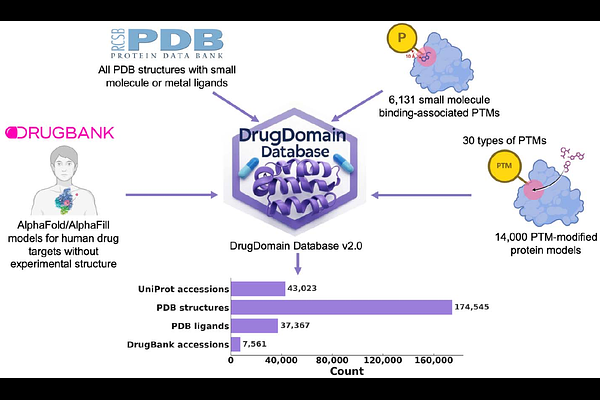
AbstractProteins carry out essential cellular functions - signaling, metabolism, transport - through the specific interaction of small molecules and drugs within their three-dimensional structural domains. Protein domains are conserved folding units that, when combined, drive evolutionary progress. The Evolutionary Classification Of protein Domains (ECOD) places domains into a hierarchy explicitly built around distant evolutionary relationships, enabling the detection of remote homologs across the proteomes. Yet no single resource has systematically mapped domain-ligand interactions at the structural level. To fill this gap, we introduce DrugDomain v2.0, the first comprehensive database linking evolutionary domain classifications (ECOD) to ligand binding events across the entire Protein Data Bank. We also leverage AI-driven predictions from AlphaFold to extend domain-ligand annotations to human drug targets lacking experimental structures. DrugDomain v2.0 catalogs interactions with over 37,000 PDB ligands and 7,560 DrugBank molecules, integrates 6,000+ small-molecule-associated post-translational modifications, and provides context for 14,000+ PTM-modified human protein models featuring docked ligands. The database encompasses 43,023 unique UniProt accessions and 174,545 PDB structures. The DrugDomain data is available online: https://drugdomain.cs.ucf.edu/ and https://github.com/kirmedvedev/DrugDomain.