Evaluating microbial network inference methods: Moving beyond synthetic data with reproducibility-driven benchmarks
Evaluating microbial network inference methods: Moving beyond synthetic data with reproducibility-driven benchmarks
Ghaeli, Z.; Aghdam, R.; Eslahchi, C.
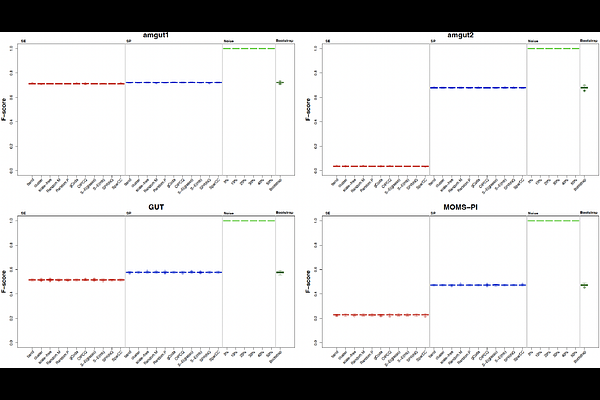
AbstractMicrobial network inference is an essential approach for revealing complex interactions within microbial communities. However, the lack of experimentally validated gold standards presents a significant obstacle in evaluating the accuracy and biological relevance of inferred networks. This study delivers a comprehensive comparative assessment of six widely used microbial network inference algorithms--gCoda, OIPCQ, S-E(glasso), S-E(mb), SPRING, and SparCC--using four diverse real-world microbiome datasets alongside multiple types of generated data, including synthetic, noisy, and bootstrap-derived datasets. Our evaluation framework extends beyond conventional synthetic benchmarking by emphasizing reproducibility-focused assessments grounded in biologically realistic perturbations. We show that bootstrap resampling and low-level noisy datasets (<=10% perturbation) effectively preserve key statistical properties of real microbiome data, such as diversity indices, abundance distributions, and sparsity patterns. Conversely, synthetic datasets generated via the widely used SPIEC-EASI method exhibit substantial divergence from real data across these metrics. We find that while SparCC demonstrates superior robustness across varying data conditions, other methods tend to produce inflated performance metrics when evaluated on unrealistic synthetic networks. Notably, several algorithms fail to distinguish between structured and random networks, highlighting issues of structural insensitivity and the limitations of overreliance on synthetic benchmarks. To address these challenges, we propose a reproducibility-centered benchmarking framework that prioritizes real-data-derived perturbations and mandates rigorous statistical validation of synthetic datasets before their use. This work provides critical insights and practical guidance for the microbiome research community, aiming to foster more reliable and ecologically meaningful microbial network inference in the absence of a true ground truth.