Validation of Morphology-Guided Computational Enhancement for Single-Cell Resolution Spatial Transcriptomics
Validation of Morphology-Guided Computational Enhancement for Single-Cell Resolution Spatial Transcriptomics
Elbialy, A.
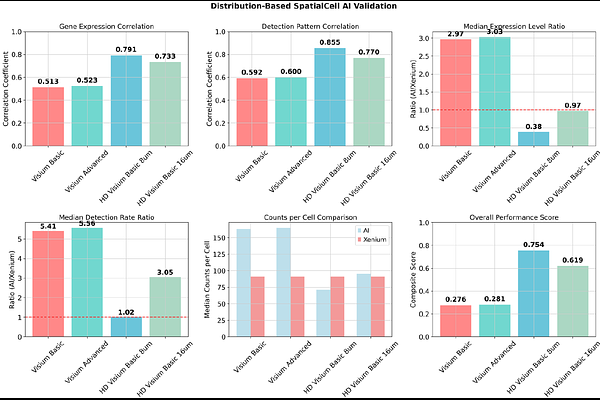
AbstractSpatial transcriptomics technologies face a fundamental trade-off between transcriptomic breadth and spatial resolution, with widely-used platforms like 10x Visium capturing multiple cells per spot, limiting single-cell insights. Current computational deconvolution methods attempt to address this limitation but uniformly suffer from reference dependency, platform effects, and complete neglect of tissue morphology. Here we present SpatialCell AI, a computational framework that achieves true single-cell resolution from spot-based spatial transcriptomics through morphology-guided computational enhancement. Unlike existing methods that rely solely on expression similarity, SpatialCell AI integrates AI-powered cell segmentation from histological images with spatial gene expression, eliminating reference requirements while leveraging tissue architecture. We rigorously validated our approach using publicly available matched colorectal cancer samples analyzed across Visium (55m), Visium HD (8m, 16m), and Xenium (single-cell ground truth). SpatialCell AI achieved strong accuracy with expression correlation of r=0.791, 7.82-fold improvement in expression accuracy, and 5.30-fold enhancement in gene detection compared to spot-based measurements. Comprehensive benchmarking against 28+ existing methods revealed that all computational approaches share identical limitations that SpatialCell AI uniquely overcomes through its morphology-first design. The framework converts standard Visium outputs from spot-level to true single-cell resolution (Cell_1, Cell_2, Cell_3...), enabling precise cellular interaction mapping and rare cell type identification previously impossible with spot-based technologies. By bridging the resolution gap between affordable spot-based platforms and expensive single-cell technologies, this approach enables broader access to single-cell resolution analysis for research and clinical applications.