OmiXAI: An Ensemble XAI Pipeline for Interpretable Deep Learning in Omics Data
OmiXAI: An Ensemble XAI Pipeline for Interpretable Deep Learning in Omics Data
Alaeva, A.; Lapteva, A.; Mikhaylovskaya, N.; Malkov, V.; Herbert, A.; Borevskiy, A.; Poptsova, M.
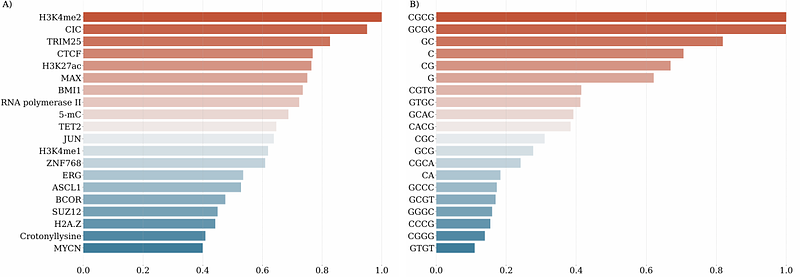
AbstractDeep learning methods have become methods of choice in the analysis of genomic data. The performance of deep learning models depends on the information available for training. A growing trend in deep learning applications involves leveraging multi-omics data spanning genomics, transcriptomics, epigenomics, proteomics, metabolomics, and other domains. When a deep learning model trained on omics data achieves high performance, the important question is to define factors that contribute to model predictive power. Explainable AI (XAI) methods can be categorized as model-aware and model-agnostic. Model-agnostic approaches, which rely on combinatorial feature perturbations to assess impact, are often computationally prohibitive for deep learning models. To address this, we developed OmiXAI, a pipeline integrating ensemble model-aware XAI methods. Our framework incorporates gradient-based techniques including Integrated Gradients, InputXGradients, Guided Backpropagation, and Deconvolution (for CNNs and GNNs) as well as Saliency Maps and GNNExplainer (specifically for GNNs). We evaluated OmiXAI on Z-DNA prediction using multi-omics features, demonstrating its efficacy through feature importance analysis and benchmarking of XAI methods. Notably, OmiXAI enabled feature engineering, reducing the critical feature set from almost 2,000 to just 50. Its modular design allows seamless integration of additional attribution methods, ensuring adaptability beyond omics to diverse problem domains. While testing the ensemble approach we benchmarked individual XAI methods and discuss their drawbacks and limitations. OmiXAI is freely available at https://github.com/aameliig/OmiXAI.