SPAED: Harnessing AlphaFold Output for Accurate Segmentation of Phage Endolysin Domains
SPAED: Harnessing AlphaFold Output for Accurate Segmentation of Phage Endolysin Domains
Boulay, A.; Cremelie, E.; Galiez, C.; Briers, Y.; Rousseau, E.; Vazquez, R.
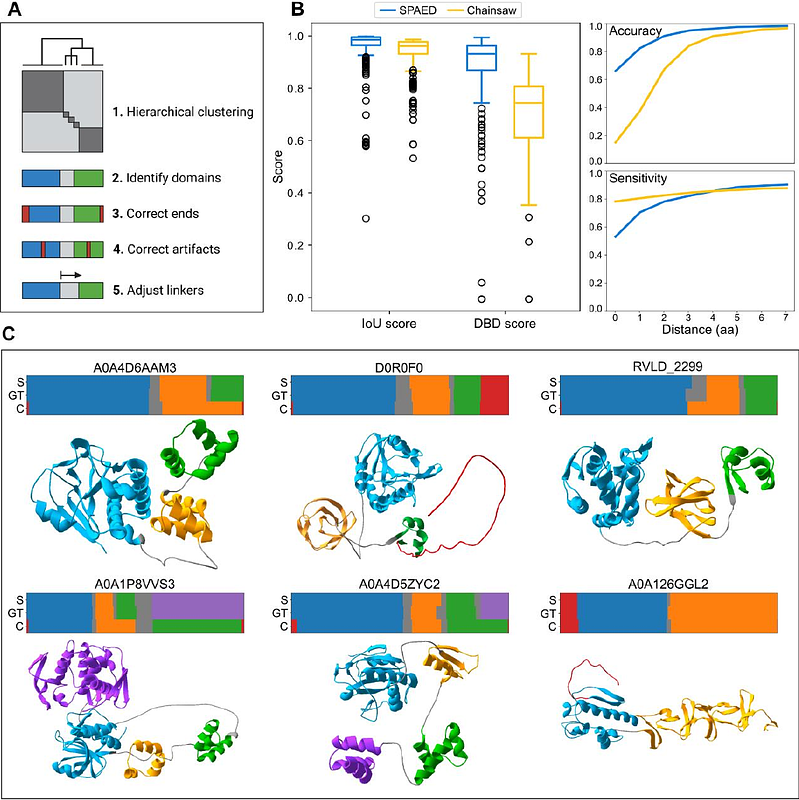
AbstractSPAED is an accessible tool for the accurate segmentation of protein domains that applies hierarchical clustering to the predicted aligned error (PAE) matrix obtained from AlphaFold predictions. It leverages information contained in the PAE matrix to better identify domain-linker boundaries and detect disordered regions. On a dataset of 376 bacteriophage endolysins (proteins that degrade the bacterial cell wall), SPAED achieves a mean intersect-over-union score of 96% and a domain-boundary-distance score of 89% compared to 94% and 70%, respectively, for the state-of-the-art tool Chainsaw. SPAED is available on the web at http://spaed.ca and available for download at https://github.com/Rousseau-Team/spaed.