A global survey of intramolecular isopeptide bonds
A global survey of intramolecular isopeptide bonds
Costa, F.; Riziotis, I.; Andreeva, A.; Kalwan, D.; de Jong, J.; Hinchliffe, P.; Parmeggiani, F.; Race, P. R.; Burston, S. G.; Bateman, A.; Barringer, R.
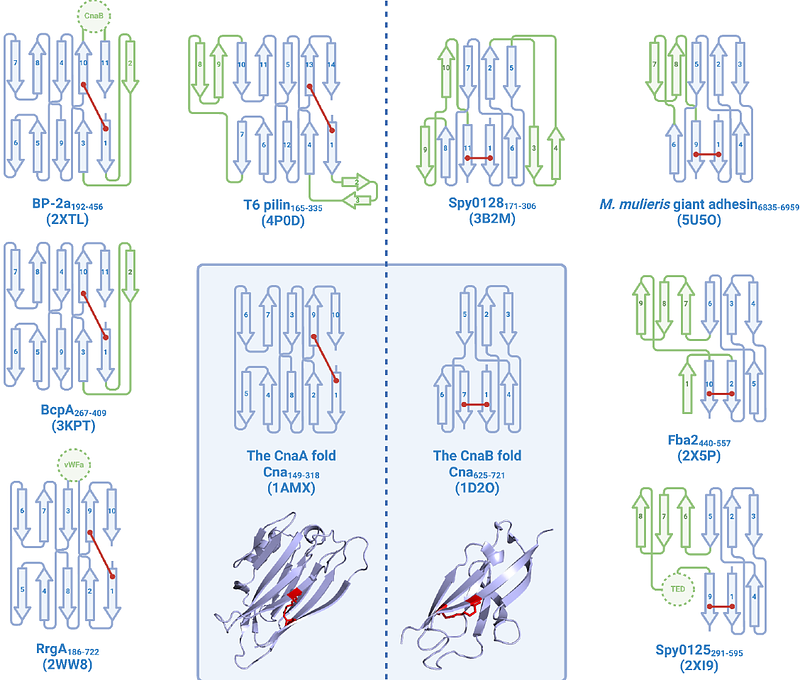
AbstractMany protein domains harbour covalent intramolecular bonds that enhance their stability and resistance to thermal, mechanical and proteolytic insults. Intramolecular isopeptide bonds represent one such covalent interaction, yet their distribution across protein domains and organisms has been largely unexplored. Here, we sought to address this by employing a large-scale prediction of intramolecular isopeptide bonds in the AlphaFold database using the structural template-based software Isopeptor. Our findings reveal an extensive phyletic distribution in surface proteins resembling fibrillar adhesins and pilins. All identified intramolecular isopeptide bonds are found in two structurally distinct folds, CnaA-like or CnaB-like, from a relatively small set of related Pfam families, including ten novel families that we predict to contain intramolecular isopeptide bonds. One CnaA-like domain of unknown function, DUF11 (renamed here to \"CLIPPER\") is broadly distributed in cell-surface proteins from Gram-positive bacteria, Gram-negative bacteria, and archaea, and is structurally and biophysically characterised in this work. Using X-ray crystallography, we resolve a CLIPPER domain from a Gram-negative fibrillar adhesin that contains an intramolecular isopeptide bond and further demonstrate that it imparts thermostability and resistance to proteolysis. Our findings demonstrate the extensive distribution of intramolecular isopeptide bond-containing protein domains in nature, and structurally resolve the previously cryptic CLIPPER domain.