XpressO-Melanoma: An Explainable Deep Learning Model for the Prediction of BRAF V600 Mutation Status in Cutaneous Melanomas
XpressO-Melanoma: An Explainable Deep Learning Model for the Prediction of BRAF V600 Mutation Status in Cutaneous Melanomas
R Rao, V.; Phung, T.; Sukhadia, S. S.
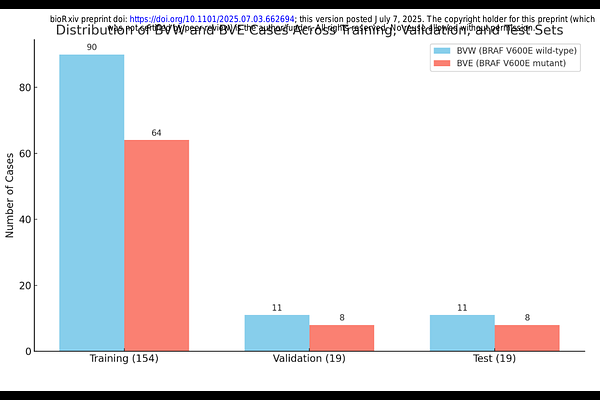
AbstractBRAF V600E mutations are critical oncogenic drivers in cutaneous melanoma, influencing treatment decisions and outcomes. However, conventional molecular assays face limitations, including tissue availability, cost, and access. To address this, we present an explainable deep learning model that predicts BRAF V600E mutation status directly from diagnostic whole-slide images (WSIs) of skin cutaneous melanoma. Using histopathological WSIs from The Cancer Genome Atlas (TCGA) and their corresponding mutation labels (BRAF wildtype vs. BRAF V600E), we trained a weakly supervised deep learning pipeline, XpressO, to identify tumor regions of interest (ROIs) predictive of BRAF mutation status. The model outputs attention heatmaps highlighting spatially relevant diagnostic features and computes a combined probability score from the top ten attention regions per WSI. These regions are further reviewed by a pathologist for biological appropriateness. On an independent test set, the model achieved an AUC of 0.79 with balanced precision and recall, correctly identifying 7 of 8 BRAF V600E mutant cases. This demonstrates the model\'s ability to capture phenotypic correlates of mutation status and highlights the potential of computational pathology in precision oncology. Our approach offers a scalable, interpretable, and cost-effective alternative to molecular testing, particularly in resource-limited settings.