Phage-Assisted Evolution of Allosteric Protein Switches
Phage-Assisted Evolution of Allosteric Protein Switches
Southern, N. T.; von Bachmann, A.-L.; Hovsepyan, A.; Griebl, M.-L.; Wolf, B.; Lemmen, N.; Kroell, A.-S.; Westermann, S.; Mathony, J.; Niopek, D.
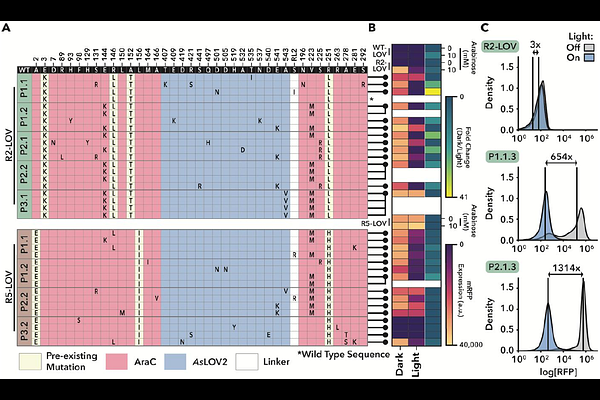
AbstractAllostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation of cell function; their engineering, however, is hampered by our limited understanding of allosteric residue networks. Here, we introduce a phage-assisted evolution platform for in vivo optimization of allosteric proteins. It applies opposing selection pressures to enhance activity and switchability of phage-encoded effectors and leverages retron-based recombineering to broadly explore fitness landscapes, covering point mutations, insertions and deletions. Applying our pipeline to the transcription factor AraC yielded optogenetic variants with light-controlled activity spanning ~1000-fold dynamic range. Long-read sequencing across selection cycles revealed adaptive trajectories and corresponding allosteric interactions. Our work facilitates phage-assisted evolution of allosteric proteins for programmable cellular control.