From Pairwise Distances to Neighborhood Preservation: Benchmarking Dimensionality Reduction Algorithms for CyTOF, scRNA-seq, and CITE-seq
From Pairwise Distances to Neighborhood Preservation: Benchmarking Dimensionality Reduction Algorithms for CyTOF, scRNA-seq, and CITE-seq
Bombina, P.; Adams, Z. B.; McGee, R. L.; Coombes, K. R.
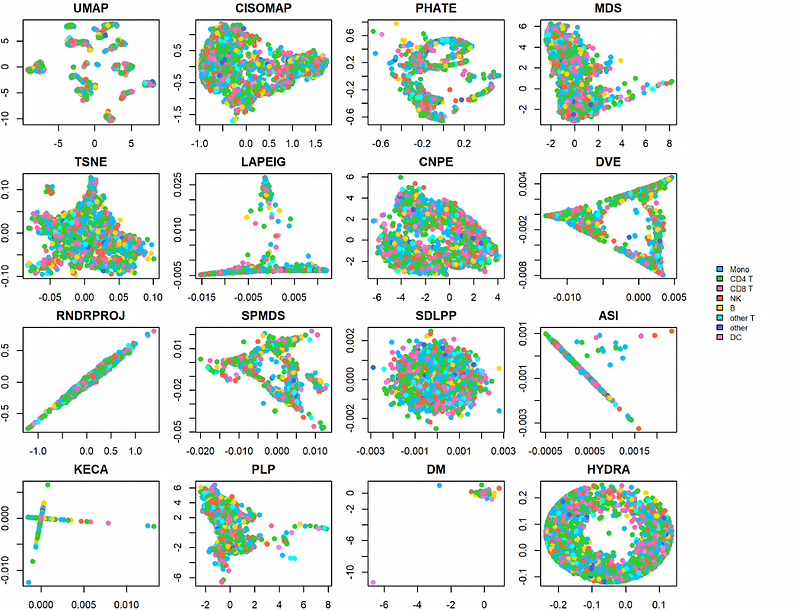
AbstractDimensionality reduction algorithms are essential tools for visualizing high-dimensional biological data, such as single-cell transcriptomics, mass cytometry by time of flight, and cellular indexing of transcriptomes and epitopes. These algorithms map complex, high dimensional data into lower dimensions to reveal underlying structures and patterns. Despite the popularity of dimensionality reduction methods like t-SNE and UMAP, concerns have arisen regarding their ability to preserve critical aspects of high-dimensional data and their sensitivity to user-defined parameters. This study aims to evaluate the impact of extreme dimension reduction from hundreds or thousands of dimensions to just two dimensions, highlighting the resulting distortions to deeply understand their implications. Given the significance of dimensionality reduction in biological research, careful evaluation of these methods is necessary to ensure reliable and meaningful results. In this paper, we present a comprehensive evaluation of 16 dimensionality reduction methods. Our evaluation addresses several key factors, such as the preservation of pairwise distances and local neighborhood relationships between the original high-dimensional space and the low-dimensional projections.