EndoGenius: Enabling comprehensive identification and quantitation of endogenous peptides
EndoGenius: Enabling comprehensive identification and quantitation of endogenous peptides
Fields, L.; Dang, T. C.; Gray, M. D.; Protya, S. S.; Li, l.
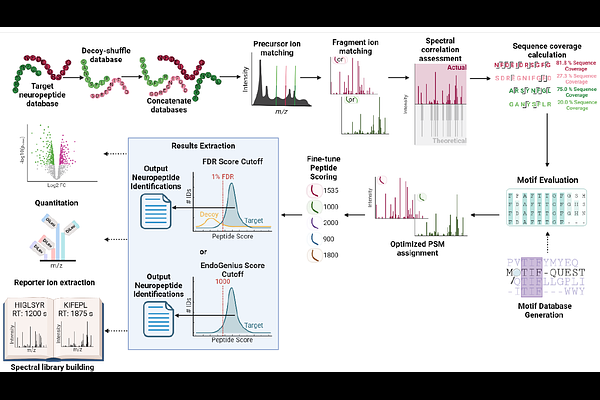
AbstractThe investigation of endogenous peptides, specifically with respect to neuropeptides, from mass spectrometry data is rife with bioinformatics bottlenecks, stemming from the low in vivo abundance of these analytes, increased susceptibility to degradation, and an immense search space of possible peptides. To address this, we present EndoGenius in its expanded form, strategically designed to optimize the searching for these endogenous peptides complemented with a pipeline designed for tasks including quantitation, spectral library building, motif extraction, and usage with data-independent acquisition workflows. EndoGenius is released as an open-source software package under an MIT License. The EndoGenius package with a user interface can be installed from https://www.lilabs.org/resources. The source code for EndoGenius can be accessed at https://github.com/lingjunli-research/EndoGenius-v2.0.