GAME: Genomic API for Model Evaluation
GAME: Genomic API for Model Evaluation
Luthra, I.; Priyadarshi, S.; Guo, R.; Mahieu, L.; Kempynck, N.; Dooley, D.; Penzar, D.; Vorontsov, I.; Sheng, Y.; Tu, X.; Klie, A.; Drusinsky, S.; Floren, A.; Armand, E.; Alasoo, K.; Seelig, G.; Tewhey, R.; Koo, P.; Agarwal, V.; Gosai, S.; Pinello, L.; White, M. A.; Lal, A.; Zeitlinger, J.; Pollard, K. S.; Libbrecht, M.; Carter, H.; Mostafavi, S.; Kulakovskiy, I.; Hsiao, W.; Aerts, S.; Zhou, J.; de Boer, C. G.
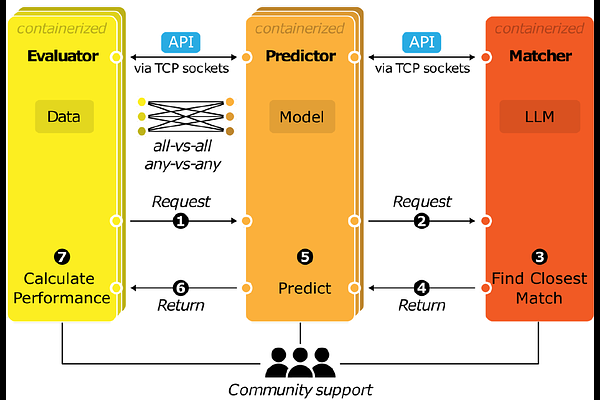
AbstractThe rapid expansion of genomics datasets and the application of machine learning has produced sequence-to-activity genomics models with ever-expanding capabilities. However, benchmarking these models on practical applications has been challenging because models are evaluated in heterogenous and ad hoc ways. To address this, we have created GAME, a system for large-scale, community-led standardized model benchmarking on diverse evaluation tasks. GAME uses Application Programming Interfaces (APIs) to communicate between containerized pre-trained models and benchmarking tasks, enabling uniform model evaluations and cross-platform compatibility. We also developed a Matcher module powered by a large language model (LLM) to automate ambiguous task alignment between prediction requests and tasks models can perform. The community can easily contribute GAME modules, leading to an ever-expanding and evolving set of models and benchmarks. GAME will accelerate genomics research by illuminating the best models for a given task, motivating novel benchmarks, and providing a nuanced understanding of model abilities.