ANABAG: Annotated Antibody Antigen dataset with unique features for Antibody Engineering Applications
ANABAG: Annotated Antibody Antigen dataset with unique features for Antibody Engineering Applications
Grandguillaume, I.; Etchebest, c.; Barroso da Silva, F. L.
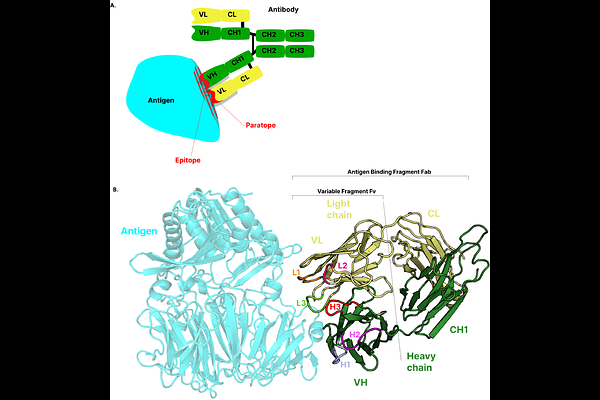
AbstractThe analysis and prediction of antibody-antigen (Ab-Ag) interactions often overlook critical structural features such as glycosylation, physical chemical conditions like pH and salt concentration, as well as the lack of standardized criteria for selecting complexes based on structural properties and sequence identity. Common practices in dataset construction rely on removing redundancy using sequence identity thresholds, which can inadvertently exclude complexes with alternative binding modes that share identical sequences. To enable more precise Ab-Ag modeling and antibody engineering, it is essential to incorporate richer structural and physical information into both physics based and machine learning models. To address these limitations, we present ANABAG, a new curated dataset of Ab-Ag complexes annotated at the residue level with UniProt sequence information and enriched with a wide range of structural and physicochemical features. The dataset allows flexible filtering of complexes using a variety of descriptors available at both the complex and residue levels. Selected features are ready to use in machine learning workflows, while the structural files are compatible with antibody design and docking pipelines like Rosetta or Haddock. The complete dataset is available on Zenodo, and all accompanying scripts and usage documentation can be accessed via GitHub at https://github.com/DSIMB/anabag-handler.git.