Learning copy number dependent variation in single tumour cell transcriptomes with deep generative models
Learning copy number dependent variation in single tumour cell transcriptomes with deep generative models
Selega, A.; Maan, H.; Yu, C.; Tan, T. J.; Campbell, K. R.
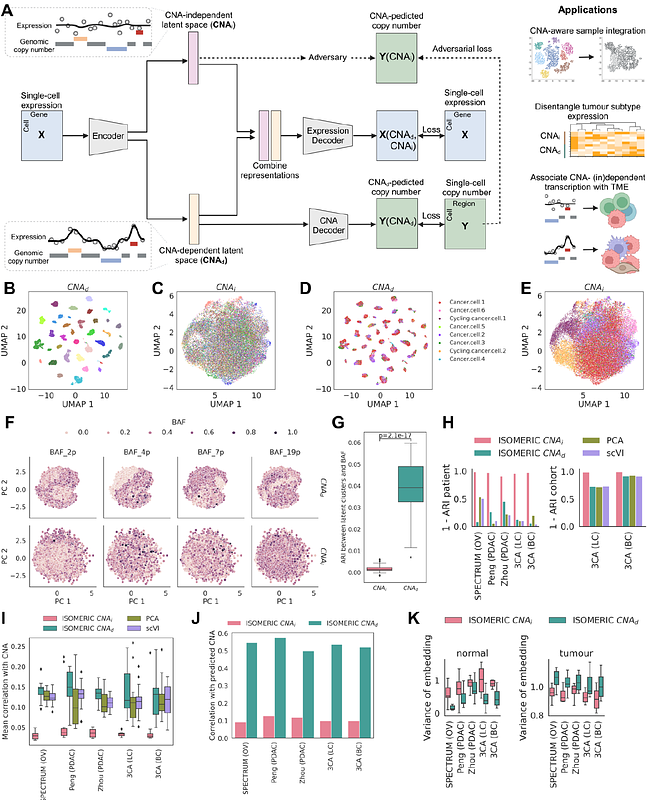
AbstractLarge-scale copy number aberrations are a hallmark of many cancers, showing widespread associations with tumour cell phenotypes, therapy resistance, and patient outcomes. Several existing methods can infer copy number profiles from single-cell RNA-sequencing data. However, there is currently no methodology to separate the effects of the copy number state on single tumour cell phenotypes from the copy number-independent variation. We present ISOMERIC, an unsupervised deep learning framework to learn disentangled representations of single-cell expression data that are dependent on or independent of copy number profiles. We apply ISOMERIC to multiple cancer types and examine how copy number shapes transcriptional states. We find distinct associations between copy number-dependent and independent variation and clinical subtypes in multiple cancer types and link such variation to tumour microenvironment phenotypes. Together, this establishes a principled framework for untangling the effects of genetic and intrinsic variation on tumour transcriptomes and the consequences on clinically meaningful phenotypes.