Statistical knockoffs improve biomarker discovery fromtranscriptomic data
Statistical knockoffs improve biomarker discovery fromtranscriptomic data
CARTIER, J.; LAGOAS, J.; FERMANIAN, A.; Azencott, C.-A.; MASSIP, F.
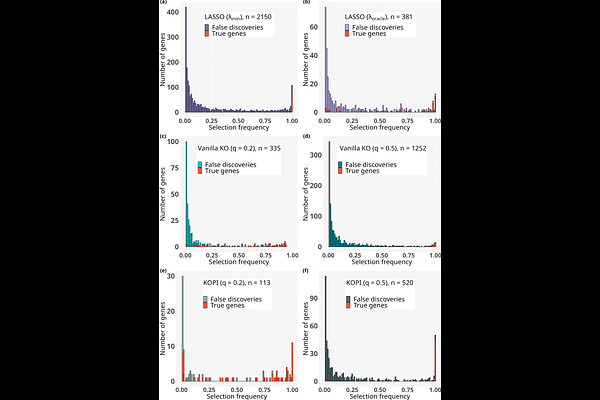
AbstractAdvances in sequencing technologies have enabled the generation of large amounts of data, offering new possibilities to identify relationships between biological units (e.g. genes) and phenotypic traits (e.g. disease outcomes). Yet, identifying these associations using variable selection methods remains challenging due to the high dimension and the correlation structure of the data. To address these challenges, we study the applicability of the knockoff (KO) procedure. Introduced by Barber and Candès in 2015, the KO variable selection procedure has shown promising results on real biological data, such as Genome Wide Association Studies. This method seeks to identify the truly important predictors by overcoming the correlation structure between variables while controlling the false discovery rate. Here, we study the applicability of the KO procedure on transcriptomic data in a classification setting. We conduct an extensive simulation study using real transcriptomic data to evaluate the performance of the KO framework in the context of high-dimensional classification. We find that the KO framework outperforms widely used variable selection models, and that using KO aggregation to mitigate the effect of KO stochasticity improves stability while maintaining the same power. Finally applied to three real transcriptomic datasets, the KO framework made very few discoveries, highlighting its conservative nature and suggesting that other methods may substantially overestimate the number of relevant features.