CAPTAIN: A multimodal foundation model pretrained on co-assayed single-cell RNA and protein
CAPTAIN: A multimodal foundation model pretrained on co-assayed single-cell RNA and protein
Ji, B.-Y.; Hu, T.-T.; Wang, J.-W.; Liu, M.-M.; Xu, L.-W.; Zhang, Q.-H.; Zhong, S.-Y.; Qiao, L.-B.; Zhang, Y.; Peng, S.-L.; Yu, F.-L.
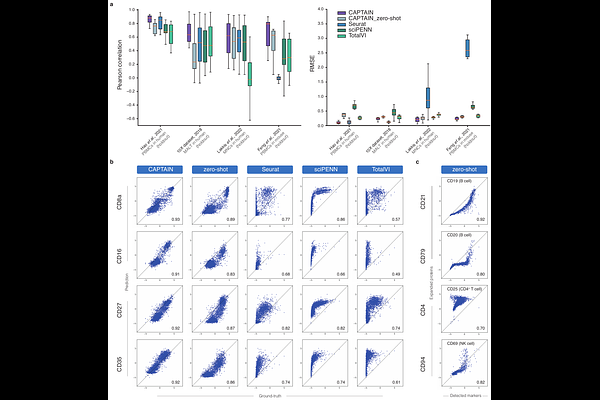
AbstractProteins act as the terminal effectors of cellular function, encoding the phenotypic consequences of genomic and transcriptomic programs. Although transcriptomic profiles serve as accessible proxies, they remain incomplete surrogates for the proteomic landscape that ultimately defines cellular phenotypes. Current single-cell foundation models, however, are trained exclusively on transcriptomes, resulting in biased and partial characterizations of cellular states. To address this limitation, we introduce CAPTAIN, a multimodal foundational model pretrained on over four million single cells with concurrently measured transcriptomes and a curated repertoire of 382 surface proteins across diverse human and mouse tissues. Our results show that CAPTAIN learns unified multimodal representations by modeling cross-modality dependencies and capturing the diversity of cellular states across complex biological contexts. CAPTAIN generalizes robustly across both fine-tuning and zero-shot settings, excelling in core downstream tasks such as protein imputation and expansion, cell type annotation, and batch harmonization. Beyond improved accuracy in multi-omics integration, CAPTAIN uncovers previously inaccessible mechanisms of protein-driven intercellular dynamics, including immune interaction patterns linked to COVID-19 severity. CAPTAIN establishes a new paradigm for multimodal single-cell modeling, laying the foundation for comprehensive cellular understanding and virtual cell construction.