Telomerase RNA gene duplications drive telomeric repeatdiversity and evolution in Andrena bees
Telomerase RNA gene duplications drive telomeric repeatdiversity and evolution in Andrena bees
Klapproth, C.; Israel, E.; Ahmed, A.; Remmel, A.; Kaether, K.-K.; Reinhardt, F.; Chen, J. J. L.; Prohaska, S. J.; Steidle, J.; Lemke, S.; Stadler, P. F.; Findeiss, S.
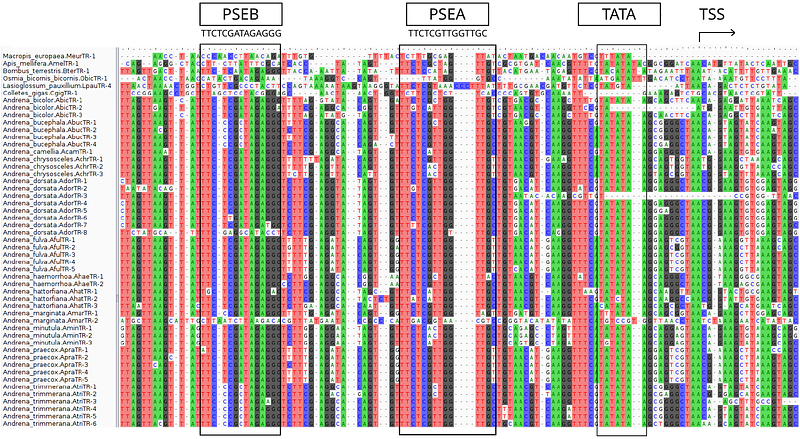
AbstractMost organisms in the animal kingdom require a non-coding telomerease RNA (TR) in conjunction with the telomerase reverse transcriptase (TERT) to add telomere tandem repeats to chromosome ends to genomic instability. Recent studies reported an extensive diversity in the sequence of telomeric repeats in some insect species. Our investigation of TR genes in the Andrena genus provides convincing evidence for the presence of multiple TR gene copies with different template sequences for synthesis of distinct telomeric repeat sequences in several species. In this study we describe the structure, genomic coordinates and abundance of these TR genes, and correlate our findings with the levels of tandem repeats found in DNAseq data. Based on an analysis of the synthenic context of these newly predicted TR genes, we show evidence for the existence of multiple TR paralogs that diverged during the evolution of the genus Andrena. To our knowledge this is the first time such a phenomenon is observed in animals, although recently reported for plants. Interestingly, the comprehensive annotation of all TERT genes found in yet unannotated Andrena species shows no corresponding evolutionary changes in related TERT proteins encoded by a single copy gene. Our study suggests an evolutionary mechanism for diversification of telomeric repeat sequences in certain insect species through telomerase RNA gene duplication.