A genome-scale CRISPR deletion screen in Chinese Hamster Ovary cells reveals essential regions of the coding and non-coding genome
A genome-scale CRISPR deletion screen in Chinese Hamster Ovary cells reveals essential regions of the coding and non-coding genome
De Marco, F.; Sebastian, I. R.; Napoleone, A.; Molin, A.; Riedl, M.; Bydlinski, N.; Motheramgari, K.; Hussein, M.; Kramer, L.; Kelly, T.; Jostock, T.; Borth, N.
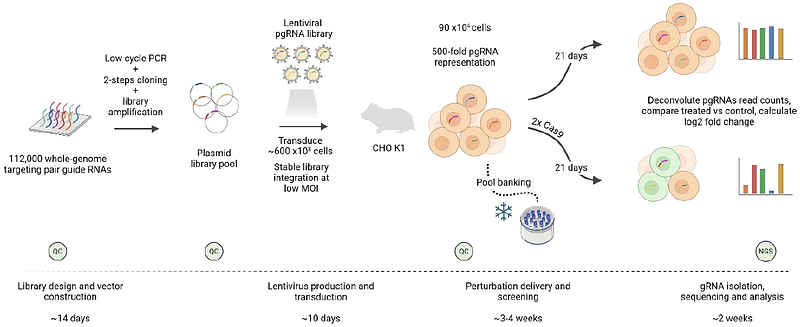
AbstractThe biopharmaceutical sector relies on CHO cells to investigate biological processes and as the preferred host for production of biotherapeutics. Simultaneously, advancements in CHO cell genome assembly have provided insights for developing sophisticated genetic engineering strategies. While the majority of these efforts have focused on coding genes, with some interest in transcribed non-coding RNAs (e.g., microRNAs and lncRNAs), there remains a lack of genome-wide systematic studies that precisely examine the remaining 90% of the genome. This unannotated \"dark matter\" includes regulatory elements and other, poorly understood or characterized functionality of the genome that may be potentially critical for cell survival. In this study, we deployed a genome-scale CRISPR screening platform with 112,272 paired guide RNAs targeting 14,034 genomic regions for complete deletion of 150 kb long sections. This platform enabled the execution of a negative screen that selectively identified dying cells to determine regions essential for cell survival. By using paired gRNAs, we overcame the intrinsic limitations of traditional frameshift strategies, which will likely have little or no effect on the non-coding genome. This study revealed 427 regions essential for CHO survival, many of which currently lack gene annotation or known function. For these regions we present annotation status, transcriptional activity as well as annotated chromatin states such as enhancers. Selected regions, specifically those that were negative for all the above, were individually deleted for confirmation. This work sheds a novel light on a substantial part of the mammalian genome which is traditionally difficult to investigate and therefore, neglected.