Clade-wide proteome analysis shows widespread non-canonical Dcr proteins in fungi.
Clade-wide proteome analysis shows widespread non-canonical Dcr proteins in fungi.
Melet, L.; Canan, J.; Villalobos, P.; Vidal-Veuthey, B.; Gonzalez-Toro, F.; Orellana, I.; Rivas-Pardo, J. A.; Cardenas, J. P.; Moraga, C.; Castro-Fernandez, V.; Johnson, N. R.; Vidal, E. A.
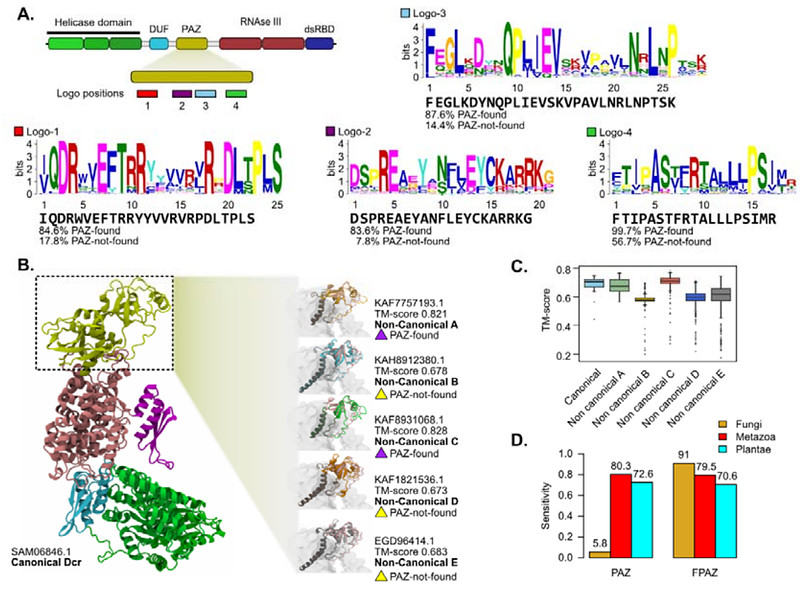
AbstractDicer (Dcr) proteins are essential components of RNA interference (RNAi) pathways, yet their evolutionary diversity and structural variation in fungi remain poorly understood. In this work, we surveyed 1,593 fungal proteomes spanning eight phyla, identifying 2,917 putative Dcr proteins. We found a predominance of non-canonical Dcr types, in which one or more domains such as Piwi/Argonaute/Zwille (PAZ), Helicase or double-stranded RNA binding domain failed to be detected, suggesting loss or sequence divergence. Phylogenetic analyses showed lineage-specific patterns of domain retention and loss, with canonical Dcrs restricted to members of the Mucoromycota and Basidiomycota. Despite high sequence divergence, AlphaFold-based structural predictions revealed conserved L-shaped architectures and PAZ-like folds in divergent Dcrs. Using Molecular simulations and electrostatic analysis, we characterized a divergent fungal PAZ found in non-canonical Dcrs, which maintains conserved motifs and RNA-binding surface properties found in canonical PAZ domains. Our findings highlight the evolutionary plasticity and functional relevance of non-canonical RNAi machinery in fungi.