Navigating parasite antigen genetic diversity in the design of Plasmodium vivax serological exposure markers for malaria
Navigating parasite antigen genetic diversity in the design of Plasmodium vivax serological exposure markers for malaria
Bareng, P.; Wu, K. W.; Takashima, E.; Argyropoulos, D.; Smith, L.; Naung, M.; Mazhari, R.; Schoffer, K.; Kiernan-Walker, N.; Abraham, A.; Lamont, M.; Mehra, S.; Lim, P.; Sattabongkot, J.; Monteiro, W. M.; Lacerda, M. V. G. d.; Healer, J.; Chitnis, C. E.; Tham, W.-H.; Tsuboi, T.; Mueller, I.; Barry, A. E.; Longley, R. J.
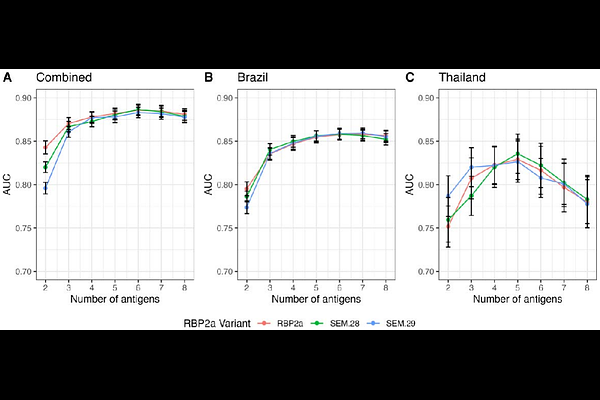
AbstractBackground: Plasmodium vivax poses a major obstacle to malaria elimination because it can lie dormant in the liver for weeks or months before reactivating and causing a relapse of infection. These dormant forms (hypnozoites) cannot be detected using standard diagnostics, but recent P. vivax exposure and by proxy, hypnozoite carriage, can be inferred using antibody-based tests (serological markers). In this study, we examined how genetic variation in P. vivax affects the utility of these antibody markers, and whether redesigned antigens could improve performance. Methods: We analysed global P. vivax genetic data to assess variation in leading serological markers. Based on this, we produced new antigen versions (haplotypes) that better reflect global sequence diversity, compared to the commonly used reference strain (Sal-1). Antibody responses against these new constructs were then tested using samples from well-characterised cohorts in Brazil and Thailand. Antibody levels were assessed in relation to how recently participants had a qPCR-detectable blood-stage P. vivax infection. We compared the ability of the haplotypes and reference constructs to correctly identify individuals infected within the prior 9-months. Findings: Extensive genetic diversity was identified in two P. vivax antigens, DBPII and MSP5. Several antigens had large numbers of circulating haplotypes globally, with the percentage with similar sequence identity to the reference Sal-1 ranging from 0.4% (MSP5) to 99% (S16). Two antigens exhibited strong differences in immunogenicity by region and construct (RBP2a and DBPII). However, for most proteins (5 out of 8), these differences had little impact on the accuracy of identifying recent exposure. In cases where performance was affected (e.g. RBP2a), this could be overcome by adding multiple antigens into the classification model. Interpretation: Even highly diverse antigens can be effective serological exposure markers. Our findings highlight the importance of testing the impact of genetic diversity when designing serological tests and suggest practical strategies, such as using a mix of antigens, to ensure consistent performance across regions.