Spatial transcriptomics of developing wheat seed reveals radial expression patterns in endosperm and subgenome biased expression of key genes
Spatial transcriptomics of developing wheat seed reveals radial expression patterns in endosperm and subgenome biased expression of key genes
Millsteed, T.; Kainer, D.; Sullivan, R.; Sun, X.; Li, K. L.; Mao, L.; Macdonald, A.; Henry, R. J.
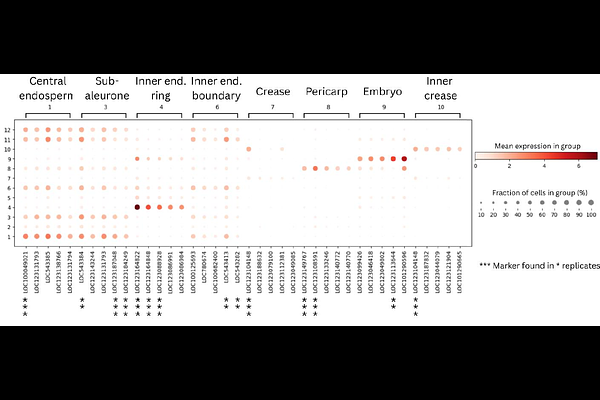
AbstractGene expression of developing seeds drives essential processes such as nutrient storage, stress tolerance and germination. However, the spatial organisation of gene expression within the complex structure of the seed remains largely unexplored. Here we report the use of the STOmics spatial transcriptomics platform to visualise spatial expression patterns in the wheat (Triticum aestivum) seed at the critical period of grain filling in mid seed development. We analysed >4,000,000 spatially resolved transcripts, achieving subcellular resolution of transcript localization across multiple tissue domains, and identified gene expression clusters linked to eight functional cellular groups. Notably, our analysis characterised four distinct clusters within the endosperm, which exhibited radial expression patterns from the inner to outer regions of the grain, and identified novel marker gene candidates for the clusters found. We further investigated known tissue-specific genes and identified subgenome biased expression between paralogs of puroindoline-B, metallothionein protein, and -amylase/subtilisin inhibitor. These findings provide new detail about gene expression across and within different functional cellular groups of the developing seed and demonstrate that spatial transcriptomics could further our understanding of subgenome differences in polyploid plants.