Bacterial species and surface structures shape gene transfer and the transcriptional landscape during early conjugation
Bacterial species and surface structures shape gene transfer and the transcriptional landscape during early conjugation
Williams, G. C.; Danne, J. C.; Crawford, S.; Homman-Ludiye, J.; Kostoulias, X.; Islam, T.; Heron, B.; Yong, M.; Gan, Y.-H.; Srikhanta, Y.; Lyras, D.
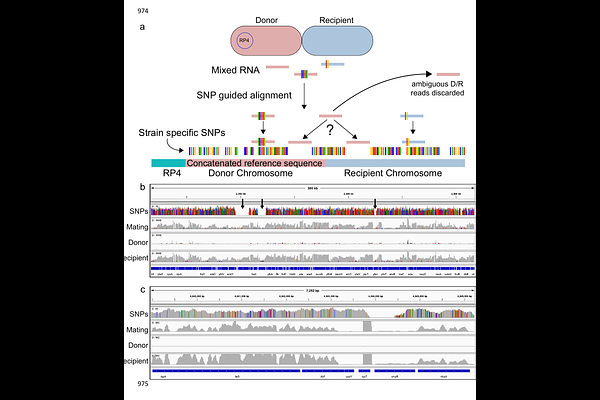
AbstractConjugative plasmids drive bacterial evolution and niche adaptation. Their carriage often corresponds with changes to the host transcriptome, but it is unknown during which stage of the conjugation process these transcriptional changes are initiated, and how gene transfer is modulated as they occur. This is because they are normally studied after their full acquisition by conjugation has already occurred, not during the process. Here, active RP4 conjugation revealed that the transcriptional response to plasmid acquisition is almost immediate and is host and surface factor dependent. Responses included activation of non-SOS stress pathways, motility, exopolysaccharide production, anaerobic respiration, and metabolic adaptation. These responses were not a barrier to conjugation, indicating that their significance relates to host homeostasis rather than gene transfer. The recipient could prevent these responses by blocking conjugation through capsule expression. RP4 expression was also inhibited by recipient capsule, since capsule physically blocked donor-recipient contact. Unexpectedly, within a population, RP4 was found to exploit single cell variation in recipient capsule thickness and to successfully conjugate with recipient bacteria with reduced capsule. These results show a novel interplay between plasmid and surface architecture across diverse species that controls the conjugation transcriptional landscape, with important ramifications for plasmid dissemination and evolution.