Single nucleotide polymorphisms and structural variants reveal complex and variable ploidy in the amoebozoan Acanthamoeba castellanii
Single nucleotide polymorphisms and structural variants reveal complex and variable ploidy in the amoebozoan Acanthamoeba castellanii
Colp, M. J.; Archibald, J. M.
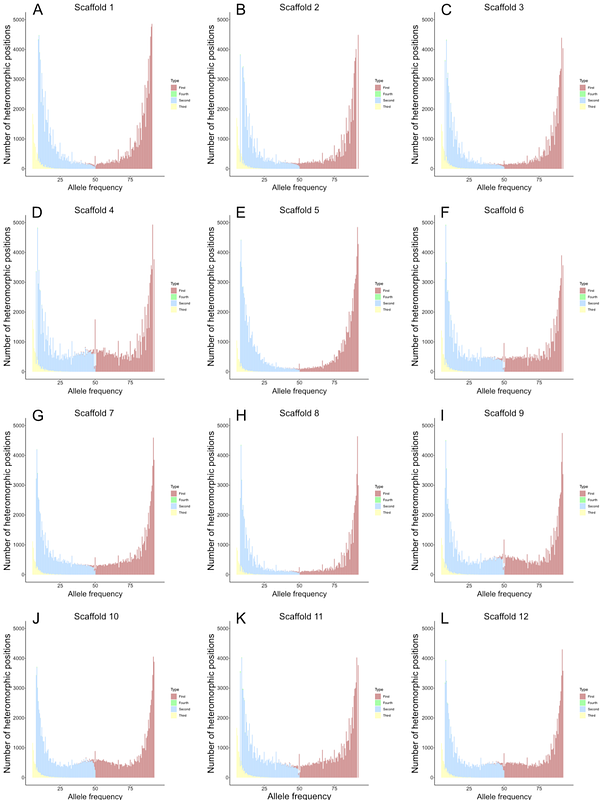
AbstractAcanthamoeba castellanii is a free-living amoeba that is emerging as a model organism for the study of eukaryotic microbiology. It is one of the most widely studied members of the Amoebozoa, and is both an important grazer in soil communities and an opportunistic human pathogen; A. castellanii is thus of evolutionary, ecological, and biomedical significance. Despite its potential as a lab workhorse, the genome biology of A. castellanii is complex and poorly understood. Polyploidy is a common feature of many amoebozoan genomes, and members of the genus Acanthamoeba are no exception; they appear to be not only polyploid, where genome copy number is inflated beyond the conventional haploid and diploid states, but also aneuploid, i.e., with inter-chromosomal copy number variation. To better understand aneuploidy in A. castellanii and how it may vary over time and between closely related strains, we analyzed nanopore and Illumina sequence datasets from several wild-type and mutant A. castellanii lines, with a focus on quantifying single nucleotide polymorphism (SNP) and structural variant allele frequencies across chromosome-scale scaffolds. Our findings suggest that intragenomic chromosome copy number is highly variable in Acanthamoeba and can change dynamically even over laboratory time scales.