Perturbational fitness analysis of CRISPR screens uncovers information-theoretic relation between gene function and selection
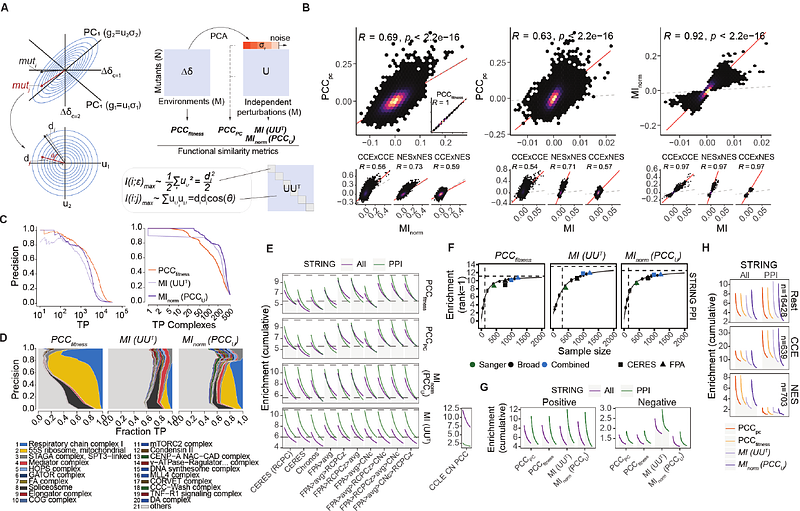
Perturbational fitness analysis of CRISPR screens uncovers information-theoretic relation between gene function and selection
Andersen, A. N.; Chica, N.; Piechaczyk, L.; Nakken, S.; Zucknick, M.; Enserink, J. M.
AbstractDespite major advances in genetic screening technology, a formal approach for quantifying gene function remains underdeveloped, thereby limiting the utility of these techniques in deciphering the complex behavior of human cells. In this study, we leverage information theory with a perturbational analysis of replicator dynamics to characterize functional drivers of selection in pooled CRISPR screens. Our approach challenges established methods for CRISPR screen analysis, while offering additional insight into selection dynamics through the Kullback-Leibler divergence (DKL) and cumulants of the fitness distribution. By modeling fluctuations in gene-fitness effects as a linear response to environmental perturbations, we derive a geometric measure for genomic information content based on a second-order approximation of the DKL. Our analysis reveals that functional information-encoded (or shared) between genes-can be quantified by analyzing the directions corresponding to maximal conditional selection within the space of decomposed gene-environment interactions. This geometric representation offers several advantages for the functional analysis of the human genome and its network architecture. Moreover, by constraining the space to cell-type-specific fluctuations, we uncover developmental and tissue-specific functional signatures. These findings represent significant progress in the dynamic analysis of gene function and in the functional wiring of the human genome.