A framework for integrating genomic profiles into captive breeding and reinforcement programmes: A case study on captive saker falcons
A framework for integrating genomic profiles into captive breeding and reinforcement programmes: A case study on captive saker falcons
Hoareau, T. B.; Barbosa, A.; Velkeneers, X.; Leveque, G.; Lesobre, L.
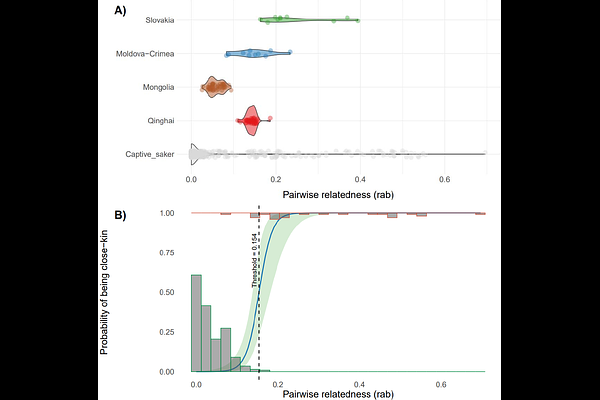
AbstractEx-situ conservation is crucial for preserving endangered species by breeding to preserve genetic diversity and provide surplus for translocation, thereby supporting in-situ conservation and enhancing wild populations. Genomic tools can assist breeding strategies and ensure long-term success of ex-situ conservation efforts by assessing genetic introgression, determining genetic origin and status, and inferring genetic relatedness of potential founders. This study aims to develop a comprehensive genomic approach for assessing the genetic profiles of candidate founders for ex-situ breeding, with the goal of releasing surplus individuals while using the endangered Saker Falcons (Falco cherrug) as a study model. Genetic clustering of 31 captive sakers revealed both diverse origins, some matching wild Asian individuals (Mongolia), and a lineage (Group III) divergent from wild populations. Comparative analyses detected hybridisation signals in 61.3% of individuals, including three with gyrfalcon (F. rusticolus) introgression and Group III\'s pronounced divergence indicating past interbreeding with an unknown falcon species. All captive birds exhibited severe inbreeding (FROH = 0.352), far exceeding wild population levels (FROH = 0.131). Using the partial pedigree data of the captive sakers, we established a genetic relatedness threshold of 0.154 (95% CI: 0.096-0.211) to identify cases of related dyads (both full and half-siblings). At this threshold, 18.3% of captive dyads showed relatedness, with asymmetric genetic contributions between pairs, reflecting a functionally small breeding flock. To avoid risks from releasing admixed or inbred individuals, we recommend excluding introgressed birds, strategically pairing purebreds, and sourcing new founders from genetically validated wild sources, especially underrepresented Central Asian lineages. Applying this genomic framework, we demonstrate its role in safeguarding genetic integrity and preventing genetic erosion in conservation breeding programmes, thereby establishing a standard for genomic-led ex-situ conservation.