High-throughput peptide-centric local stability assay extends protein-ligand identification to membrane proteins, tissues, and bacteria
High-throughput peptide-centric local stability assay extends protein-ligand identification to membrane proteins, tissues, and bacteria
Li, K.; Potel, C. M.; Becher, I.; Huettmann, N.; Garrido-Rodriguez, M.; Schwarz, J. J.; Savitski, M.
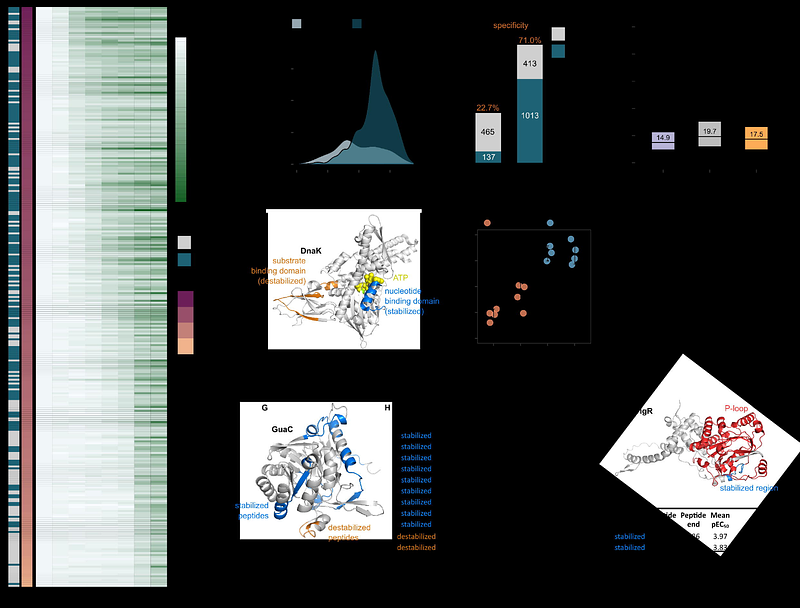
AbstractSystematic mapping of protein-ligand interactions is essential for understanding biological processes and drug mechanisms. Peptide-centric local stability assay (PELSA) is a powerful tool for detecting these interactions and localizing potential binding sites. However, its original workflow is limited in throughput, sample compatibility and accessible protein targets. Here, we introduce a high-throughput adaptation - HT-PELSA - that increases sample processing efficiency 100-fold while maintaining high sensitivity and reproducibility. HT-PELSA substantially extends the capabilities of the original method by enabling sensitive protein-ligand profiling in crude cell, tissue and bacterial lysates, making it possible to identify membrane protein targets in diverse biological systems. We demonstrate that HT-PELSA can precisely and accurately determine binding affinities of small molecule inhibitors, sensitively detect direct and allosteric ATP binding, and reveal off-target interactions of a marketed kinase inhibitor in heart tissue. By enhancing scalability, reducing costs, and enabling system-wide drug screening across a wide range of sample types, HT-PELSA - when combined with next-generation mass spectrometry - offers a powerful platform poised to accelerate both drug discovery and basic biological research.