DANSE: a pipeline for dynamic modelling of time-series multi-omics data
DANSE: a pipeline for dynamic modelling of time-series multi-omics data
Jansen Klomp, L. F.; Yan, X.; Snabel, R. R.; Veenstra, G. J. C.; Meijer, H. G. E.; Post, J. N.
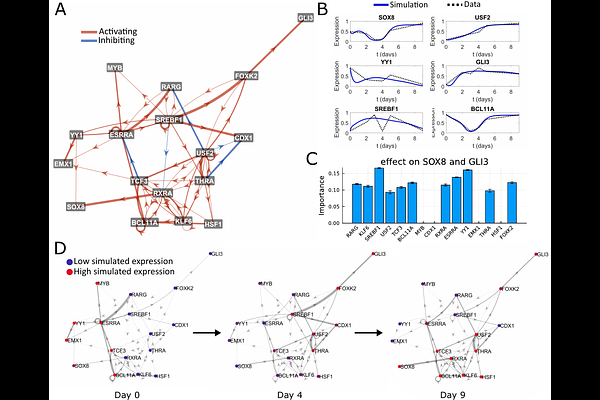
AbstractBackground Understanding time-dependent intracellular processes, such as cell differentiation, is key to developing new therapies for a wide range of diseases. Models that connect transcription factor activity to dynamic expression patterns are rare, despite the increased availability of time-series data. Results To identify key regulators of time-dependent biological processes, we present the pipeline DANSE: Dynamics inference Algorithm on Networks Specified by Enhancers. Starting from multi-omics data, our pipeline constructs a data-driven mechanistic transcription factor (TF) network and subsequently defines a dynamic model based on this TF network. The combination of a TF network and a mechanistic model allows for the identification of a small set of key transcription factors predicted to drive the modelled biological process. We showcase the result of our pipeline by applying DANSE to two different datasets that describe iPSC differentiation. Conclusions Models constructed using DANSE suggest testable hypotheses for the perturbation of gene expression, for example, knockdown or overexpression, that influence cell fate. In this way, DANSE is a powerful tool for generating novel hypotheses in a data-driven manner that take into account the dynamic nature of multi-omics time series data.