Greenhouse gas flux and microbial communities in Central Valley irrigation canals
Greenhouse gas flux and microbial communities in Central Valley irrigation canals
Kaze, M.; Tringe, S.
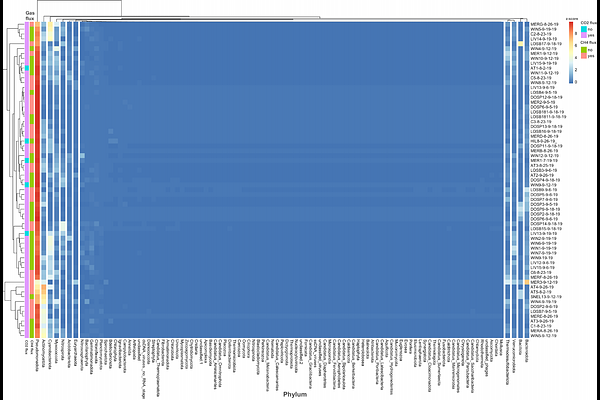
AbstractMicrobial Community Structure and Methane Production in a Large-Scale Engineered Irrigation System Climate change is of alarming and immediate concern and it is crucial to examine myriad facets contributing to the emission of greenhouse gases. Research into ecosystem microbial community structure and functional genetic diversity is essential for insight into global change and provides much-needed data for predictive models. Engineered transitional systems, such as aqueducts and networks of irrigation canals, have not been investigated for carbon flux and microbial community structure. Engineered interfaces may be acting as \'hotspots\' with impacts disproportionate to their geographic area. The Delta-Mendota aqueduct in California is 117 miles long and spans five counties; it is connected to 723 miles of irrigation canals within Merced County. This project combines field greenhouse gas measurements and culture-independent metagenomic analysis to identify the microbial communities of these two systems. This work maps the biogeographical distribution of organisms within these systems and defines their functional potential. DNA was extracted from 97 samples of sediment along 500 miles of irrigation canals in Merced County and sequenced with high-throughput whole genome sequencing. Each site was measured for methane and carbon dioxide flux with a field gas analyzer. Gas flux data was processed and visualized using multiple R packages. Metagenomic data was processed using multiple bioinformatics tools to bin and annotate genomes, normalize and quantify functional genes, and map the biogeographic distribution and phylogeny of identified microorganisms. Eighty-nine of the sample sites within the irrigation canal system demonstrated elevated carbon dioxide levels when compared to ambient air measurements (97%) and 43 sites had methane flux (44%). Results indicate a wide diversity of microorganisms present at each sampling site. Metagenomic analysis identified the microorganismal community of the sampled waterway sediments and confirmed the presence of metabolic pathways for methanogenesis and methane oxidation. Methanogenic genes were most abundant in methane producing sites. This community structure analysis reveals the potential of microbial biogeochemical activity and the gas measurements signal that greenhouse gas emission is currently occurring in these waterways. These results strongly suggest the need for monitoring and further investigation into other engineered terrestrial-aquatic interfaces.