Correlations among hydrogen bond fluctuations in the apo state are enough to reveal allosteric networks in proteins
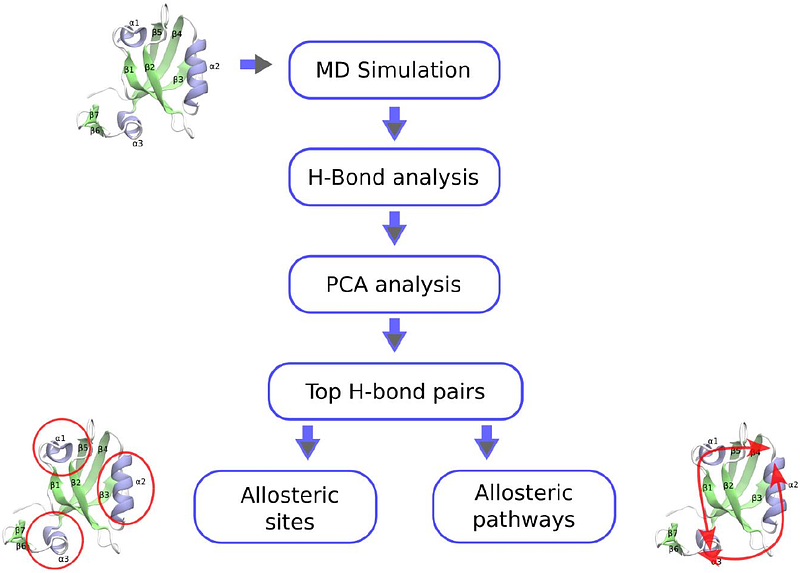
Correlations among hydrogen bond fluctuations in the apo state are enough to reveal allosteric networks in proteins
Sarkar, S.; Dhibar, S.; Jana, B.
AbstractAllostery is essential for regulating biochemical pathways, where ligand binding at one site influences enzymatic activity at a distant functional site. Identifying allosteric sites and mapping signal transduction pathways in biomolecules remain a significant challenge. Existing experimental or computational methods, in general, can identify a subset of total allosteric network. Here, we consider that in addition to providing structural stability and flexibility of proteins, hydrogen bonds inside the protein may act as conduits for long-range communication. To this end, we develop a computational framework that predicts allosteric sites and total allosteric network by analyzing correlations among hydrogen bond fluctuation from equilibrium molecular dynamics trajectory of apo state of proteins. We demonstrated that this method can accurately captures experimentally verified allosteric sites and suggest allosteric signal transduction pathways across three different proteins. Furthermore, since our predictions are derived solely from the simulation trajectory of the apo state, these findings reinforce the idea that the signature of allostery is inherently encoded in the apo state of the protein. This approach offers a useful strategy to decode allosteric network and pockets, with broad implications for drug discovery and the targeted modulation of allostery in proteins.