STIM1 transmembrane helix dimerization captured by AI-guided transition path sampling
STIM1 transmembrane helix dimerization captured by AI-guided transition path sampling
Horvath, F.; Jung, H.; Grabmayr, H.; Fahrner, M.; Romanin, C.; Hummer, G.
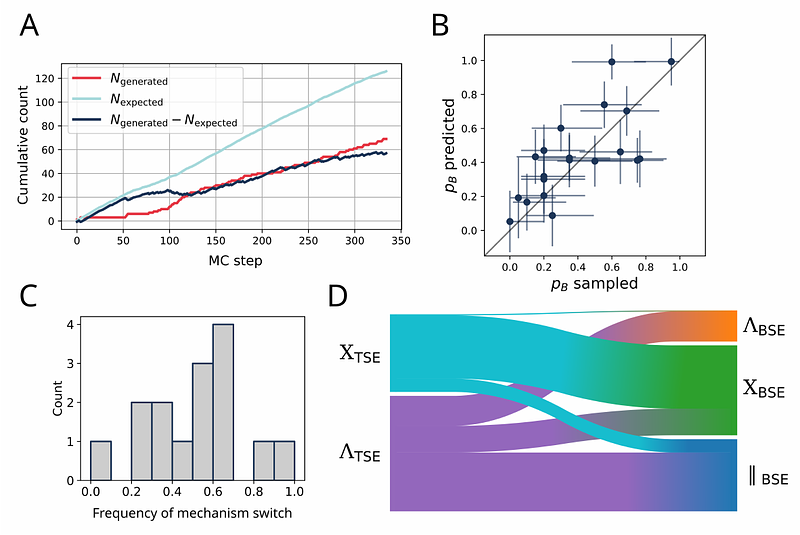
AbstractSTIM1 is a Ca2+-sensing protein in the endoplasmic reticulum (ER) membrane. The depletion of ER Ca2+ stores induces a large conformational transition of the cytosolic STIM1 C-terminus, initiated by the dimerization of the transmembrane (TM) domain. We use the AI-guided transition path sampling algorithm aimmd to extensively sample the dimerization of STIM1-TM helices in an ER-mimicking lipid bilayer. In nearly 0.5 milliseconds of all-atom molecular dynamics simulations without bias potentials, we harvest over 170 transition paths, each about 1.2 s long on average. We find that STIM1 dimerizes into three distinct and coexisting configurations, which reconciles conflicting results from earlier crosslinking studies. The dominant X-shaped bound state centers around contacts supported by the SxxxG TM interfacial motif. Mutating residues in this contact interface allows us to tune the STIM1-dimerization propensity in fluorescence experiments. From the trained model of the committor probability of dimerization, we identify the transition state ensemble for TM-helix dimerization. At the transition state, interhelical contacts in the luminal halves of the two monomers dominate, which likely enables the luminal Ca2+-sensing domain in STIM1 to condition the dimerization of the TM helices. Our work demonstrates the unique power of AI-guided simulations to sample rare and slow molecular transitions, and to produce detailed atomistic insight into the mechanism of STIM1 TM-helix dimerization as a key step in ER Ca2+-sensing.