Evolution of a fuzzy ribonucleoprotein complex in viral assembly
Evolution of a fuzzy ribonucleoprotein complex in viral assembly
Zhao, H.; Li, T.; Hassan, S. A.; Nguyen, A.; Datta, S. A.; Zhang, G.; Trent, C.; Czaja, A. M.; Wu, D.; Aronova, M. A.; Lai, K. K.; Piszczek, G.; Leapman, R. D.; Yewdell, J. W.; Schuck, P.
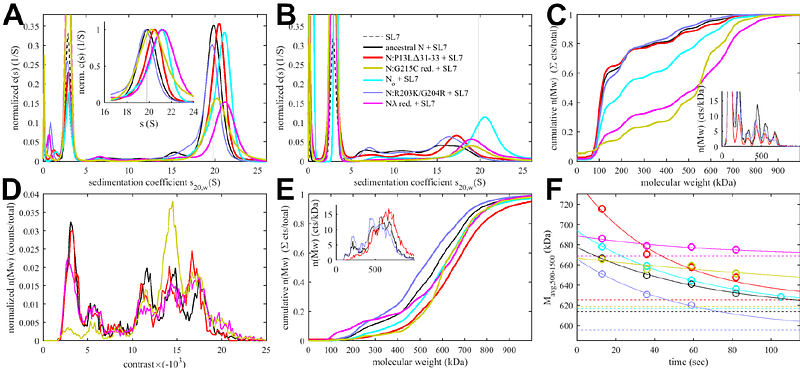
AbstractSARS-CoV-2 assembly entails condensation of viral RNA by the viral nucleocapsid (N) protein into ribonucleoprotein particles (RNPs). Lacking high-resolution structural information, biochemical and biophysical approaches have revealed their architectural principles, which involve cooperative interactions of several protein-protein and protein-RNA interfaces, initiated through oligomerization of conserved transient helices in the central disordered linker of N. Here we study the impact of defining N-protein mutations in variants of concern on RNP formation, using biophysical tools, a virus-like particle assay, and reverse genetics experiments. We find convergent evolution in independent introduction of amino acid substitutions strengthening existing binding interfaces. Furthermore, N:P13L of Omicron variants creates a self-association interface de novo, enhancing RNP assembly and increasing viral fitness. We hypothesize that the formation of polydisperse, fuzzy N-RNA clusters with distributed weak binding interfaces optimizes reversible RNA condensation while allowing for a large sequence space to be explored to support host adaptation and evolution.