Distinct chromatin regulators downmodulate meiotic axis formation and DNA break induction at chromosome ends
Distinct chromatin regulators downmodulate meiotic axis formation and DNA break induction at chromosome ends
Raghavan, A. R.; May, K.; Subramanian, V. V.; Blitzblau, H. G.; Patel, N. J.; Houseley, J.; Hochwagen, A.
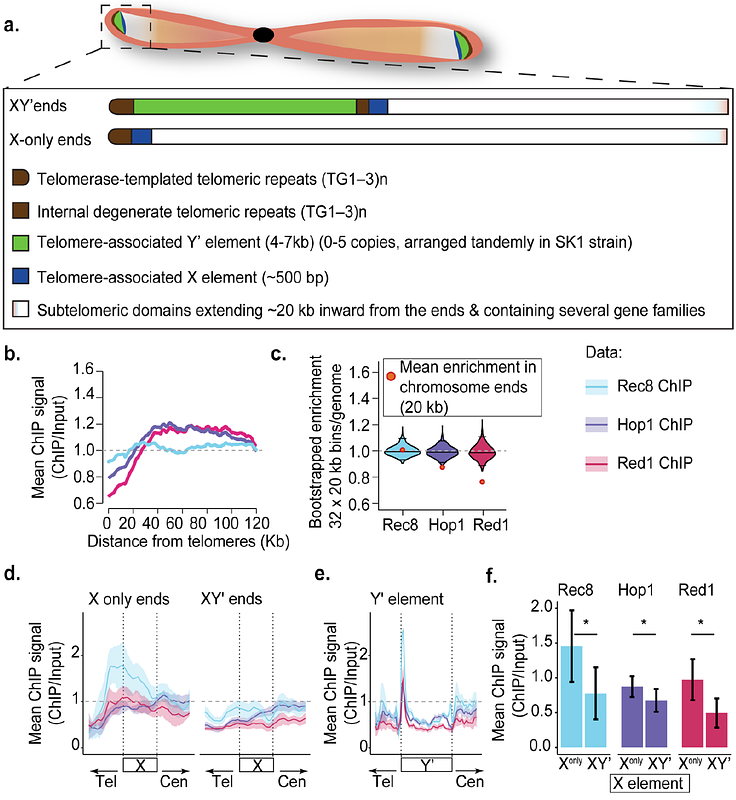
AbstractIn many organisms, meiotic crossover recombination is suppressed near the extreme ends of chromosomes. Here, we identified two chromatin modifiers, the histone methyltransferase Dot1 and the Sir silencing complex, as regulators of this process in Saccharomyces cerevisiae. We show that the recombination-promoting axis proteins Red1 and Hop1, but not the axis-associated cohesin Rec8, are significantly reduced within 20 kb of telomeres compared to the chromosome interior. Dot1, which preferentially methylates histones in the chromosome interior, is required for this pattern by directing Red1 binding toward the chromosome interior. In parallel, the Sir complex suppresses the induction of meiotic DNA double-strand breaks (DSBs) at chromosome ends. Sir-dependent DSB suppression is independent of axis deposition and occurs in a chromosome end-specific manner that mirrors the spreading and transcriptional silencing activity of the complex, suggesting that the Sir complex suppresses DSB formation by limiting the openness of promoters, the preferred sites of meiotic DSB formation. We conclude that multiple chromatin-based mechanisms collaborate to achieve a robust reduction of meiotic recombination near chromosome ends.