Case study: Genomic characteristics of the gut microbiome, Campylobacter and Salmonella genotypes in three cases of gastroenteritis co-infections
Case study: Genomic characteristics of the gut microbiome, Campylobacter and Salmonella genotypes in three cases of gastroenteritis co-infections
Djeghout, B.; Rudder, S. J.; Le Viet, T.; Elumogo, N.; Langridge, G.; Janecko, N.
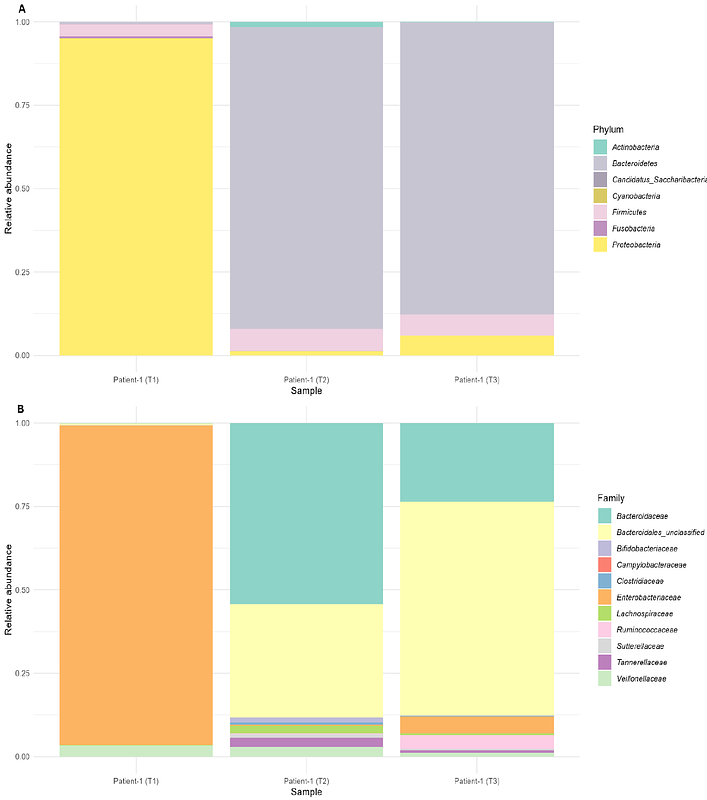
AbstractCampylobacter jejuni and Salmonella enterica co-infections in gastroenteritis cases are underexplored, particularly in relation to microbiome dysbiosis. This study uses metagenomic and culture-based approaches to investigate pathogen genotypes and stool microbiome composition. Stool samples from three patients were analysed using culture, qPCR and whole genome sequencing (WGS) of genomes and metagenomes. Strain typing and antimicrobial resistance (AMR) profiling were conducted for both pathogens. The stool microbiome structure was compared between the three patients and across three time points in one patient. C. jejuni and Salmonella enterica were of different strains and different proportions in the cases. C. jejuni sequence types were identified as ST-794, ST-50 and ST-10025, while S. enterica serovars Enteritidis ST-183 and S. Derby ST-39 were identified. C. jejuni AMR genotypes included blaOXA-447, gyrA (T86I), and tet(O). S. Enteritidis contained AMR gene mdsaA-B, and S. Derby contained mdsaA-B and fosA7 genes. Despite similarities in Bristol scales for stool composition, the three cases exhibited distinct microbiome population structures, suggesting there is no one signature microbial profile of microbiomes with a Campylobacter-Salmonella co-infection. A temporal trend towards microbial diversity was observed within a patient, shifting from Proteobacteria dominance (93.3%) during acute infection towards Bacteroidetes (89.5%) abundance post-infection, suggesting that moving towards diverse communities may play a key role in patient recovery. These findings reveal a case study of microbiome disruption for patients infected with C. jejuni and S. enterica and highlight the value of direct stool sequencing in understanding pathogen abundance in stool and microbiome dysbiosis.