Functional Insights into Dispensable Genes using Genome-Wide Loss-of-Function Burden Tests in Arabidopsis
Functional Insights into Dispensable Genes using Genome-Wide Loss-of-Function Burden Tests in Arabidopsis
Zhao, K.; Lensink, M.; Monroe, J. G.
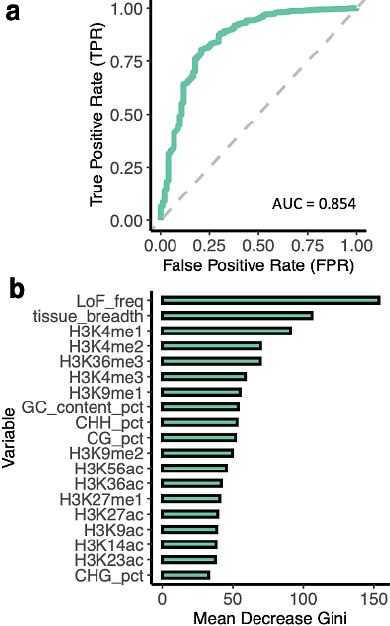
AbstractNot all genes are essential for plant survival. With the rise of pan-genomics, it is evident that certain genes can be lost without negatively affecting fitness. Naturally occurring loss-of-function (LoF) mutations provide a valuable perspective on gene dispensability, offering insights into deleterious and adaptive gene loss. In this study, we identified 91,751 naturally occurring LoF variants from publicly available Arabidopsis genome data. Our findings demonstrate that LoF-intolerant genes are enriched in essential biological functions and associated with specific histone marks linked to active transcription. In contrast, LoF-tolerant genes exhibit relaxed selective pressure and are enriched in functions related to pollen rejection and defense responses, and can be used as a proxy for dispensable genes in the pan-genome. Using a random forest model trained on histone marks, we achieved moderate success in predicting gene LoF tolerance, with an AUC of 0.717 in Arabidopsis and 0.768 in rice, and even across species. We also pioneered genome-wide LoF burden tests in Arabidopsis, collapsing independent LoF alleles into a single state to reduce allelic heterogeneity. By integrating LoF burden tests with transcriptomic data, we identified thousands of LoF-expression associations. Notably, this analysis accurately recapitulated the flowering time networks and identified FRIGIDA as a key regulator of flowering time genes. Furthermore, we found that collapsing alleles based on functional outcomes enhances association sensitivity. These results provide insight into gene dispensability and a framework for leveraging LoF mutations to study gene functions with improved association studies.