RNase H1 levels dramatically affect mitochondrial genome maintenance with little impact on nuclear R-loops in murine B cells
RNase H1 levels dramatically affect mitochondrial genome maintenance with little impact on nuclear R-loops in murine B cells
Sakhuja, K.; Hartono, S. R.; Sanz, L. A.; Ijiri, E.; Darling, C.; Dye, L.; Iben, J. R.; Chon, H.; Cerritelli, S.; Crouch, R.; Chedin, F.
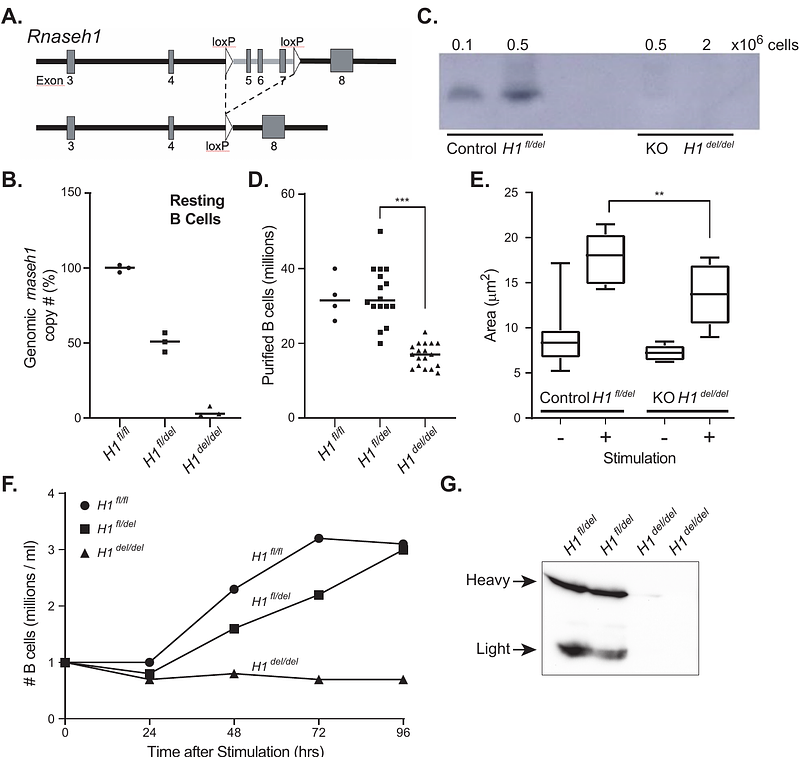
AbstractOverexpression of RNase H1, a ribonuclease that degrades RNA:DNA hybrids and R-loops, can suppress genome instability phenotypes in a range of maladaptive conditions. This has been interpreted to suggest that genotoxic co-transcriptional R-loops arise under these conditions and are resolved by RNase H1. Here, we manipulated RNase H1 levels using conditional knockout and overexpression models in primary murine B cells and mapped the resulting genomic R-loop landscapes. Rnaseh1 deletion resulted in a dramatic loss of mitochondrial replication and compromised B cell responses, consistent with a critical mitochondrial function for RNase H1. Genome-wide R-loops were, however, not significantly affected. More surprisingly, overexpressing active nuclear RNase H1 did not lead to significant reduction of R-loop levels or change their distribution. These results were confirmed using a human cell line in which active, nuclear RNase H1 can be induced. Our findings indicate that co-transcriptional R-loops are not efficiently resolved by RNase H1 under basal conditions and suggest that the identity of the RNA/DNA hybrids at the root of the genome instability phenotypes suppressed by RNase H1 may need to be re-interpreted.