Mapping the free energy landscape of K-Ras4B dimerization
Mapping the free energy landscape of K-Ras4B dimerization
Koukos, P. I.; Lesgidou, N.; Dehghani-Ghahnaviyeh, S.; Velez-Vega, C.; Duca, J. S.; Cournia, Z.
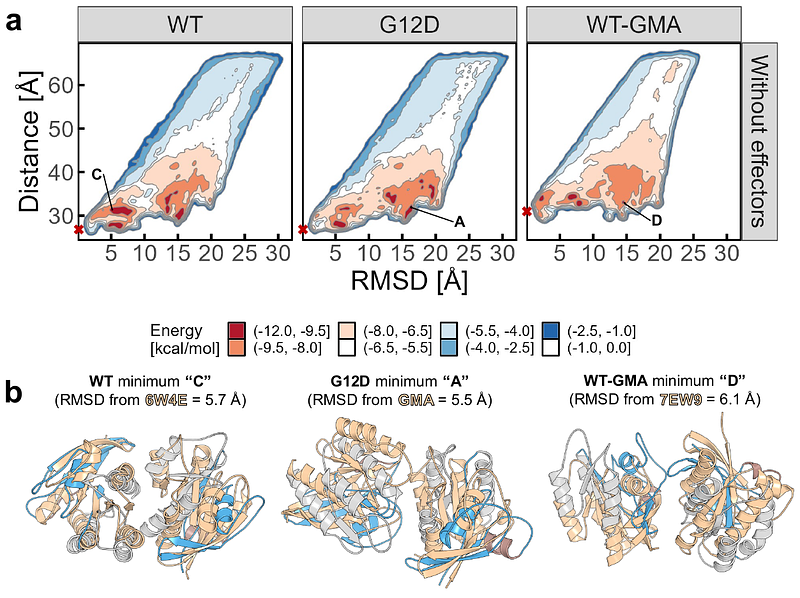
AbstractKRAS-4B regulates cellular proliferation and differentiation via its GTPase activity, and it is often mutated in human tumors. Deregulation of the MAPK/ERK pathway as a result of K-Ras4B mutations leads to uncontrolled proliferation, with the dimer/multimerization of K-Ras4B on the plasma membrane believed to be the initiating event for subsequent MAPK/ERK signaling. While K-Ras4B proteins are known to cluster on the plasma membrane, whether they associate through well-defined dimerization interfaces remains an open question. Here, we present the dimerization landscape of active, GTP-bound wild-type and G12D mutant K-Ras4B using coarse-grained unbiased and enhanced sampling molecular dynamics simulations. We recover the experimentally-reported K-Ras4B interfaces, and additionally unveil rugged free energy landscapes with many-yet uncharacterized-minima that feature c-Raf-mediated dimerization interfaces. We further explore whether wild-type or G12D K-Ras4B present different dimerization states, revealing that the G12D mutant is more likely to form diverse dimers compared to WT K-Ras4B. Our work presents evidence that K-Ras4B proteins likely interact through multifaceted interfaces that may enable controlled dimerization in different conformations from a single system, efficiently promoting nanoclustering. Although many weak, non-specific interfaces are forming, the most dominant interfaces occur with nanomolar affinity, offering a structural basis for the design of ligands able to modulate K-Ras4B dimers.