Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model Animals
Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model Animals
Wang, J.
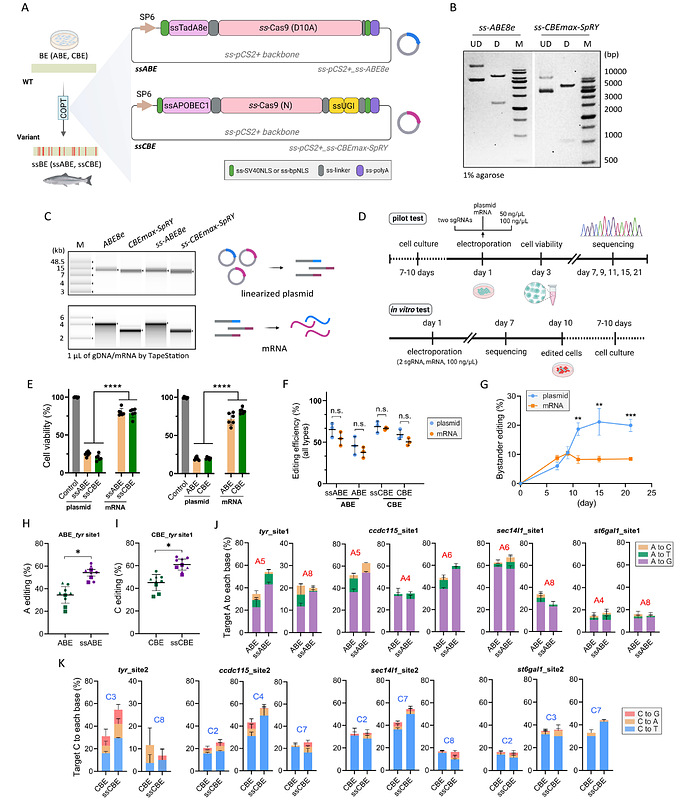
AbstractBase editors (BEs) are promising gene-editing tools for correcting point mutations in vitro and in model organisms, but their application in non-model animals remains limited. One of the major challenges is codon usage bias, as different organisms have preferences for specific codons to encode the same amino acid. Here, we engineered the constructs of variant BEs (ss-ABE8e and ss-CBE4max-SpRY) for use in a non-model animal by codon optimization and induced efficient base substitutions in vitro and in vivo. Compared to the original BEs (ABE8e and CBE4max-SpRY) derived from mammalian-cell models, our codon-optimized (COPT) BEs exhibited higher efficiency but lower bystander activity by targeting the pigment-related gene (tyr) in salmon. COPT BEs encoded by mRNA showed fewer editing patterns than plasmid-encoded BEs, mainly due to the limited lifespan of mRNA in cells. Furthermore, with these COPT editors, we were able to mutate multiple loci resulting in the complete loss of protein function in the fish genome. We ultimately generated disease-tolerant genetic variants carrying mutation-induced premature stop codons, demonstrating the value of high-resolution targeting in ss-ABE8e and ss-CBEmax-SpRY applications.