Single-molecule multimodal timing of in vivo mRNA synthesis
Single-molecule multimodal timing of in vivo mRNA synthesis
Sethi, A. J.; Guarnacci, M.; Bilal, M.; Krishnan, K. S.; Hayashi, A.; Kanchi, M.; Nojima, T.; Preiss, T.; Eyras, E.; Hayashi, R.
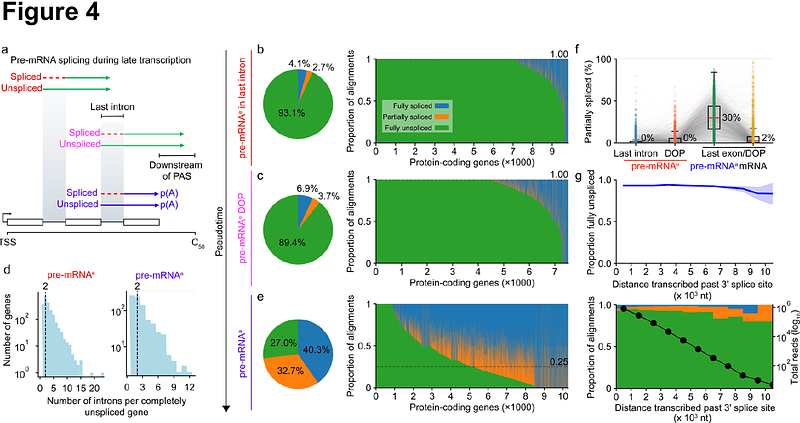
AbstractmRNA synthesis requires extensive pre-mRNA maturation, the organisation of which remains unclear. Here, we directly sequence pre-mRNA without metabolic labelling or amplification to resolve transcription and multimodal pre-mRNA processing at single-molecule resolution. Using poly(A) tail measurement, we distinguish transcriptionally engaged pre-mRNA from polyadenylated transcripts, revealing that splicing is substantially delayed compared to previous estimates. Splicing is rare for at least 10 kb behind elongating RNA polymerase II such that thousands of genes remain largely unspliced during transcription, revising the notion that splicing is predominantly co-transcriptional. Progressive splicing becomes apparent only on polyadenylated transcripts, suggesting that splicing is commonly activated after 3\' end formation. Unexpectedly, we find abundant m6A on unspliced pre-mRNA, indicating that a substantial portion of RNA methylation precedes splicing. This m6A methylation is suppressed around exon boundaries, indicating that m6A topology is established prior to exon junction complex deposition. Finally, we demonstrate that 3\' end cleavage occurs multiple kilobases behind transcription in coordination with efficient transcription termination, and further highlight recursive 3\' end formation across hundreds of genes. Conserved from human cells to mouse tissues, we illustrate a revised timeline for mRNA synthesis wherein cleavage, termination, and m6A deposition occur earlier than, or at least partially decoupled from, the bulk of splicing, reframing the sequence of early mammalian gene expression.