Elucidation of the molecular interaction network underlying full-length FUS conformational transitions and its phase separation using atomistic simulations
Elucidation of the molecular interaction network underlying full-length FUS conformational transitions and its phase separation using atomistic simulations
Weng, S. L.; Mohanty, P.; Mittal, J.
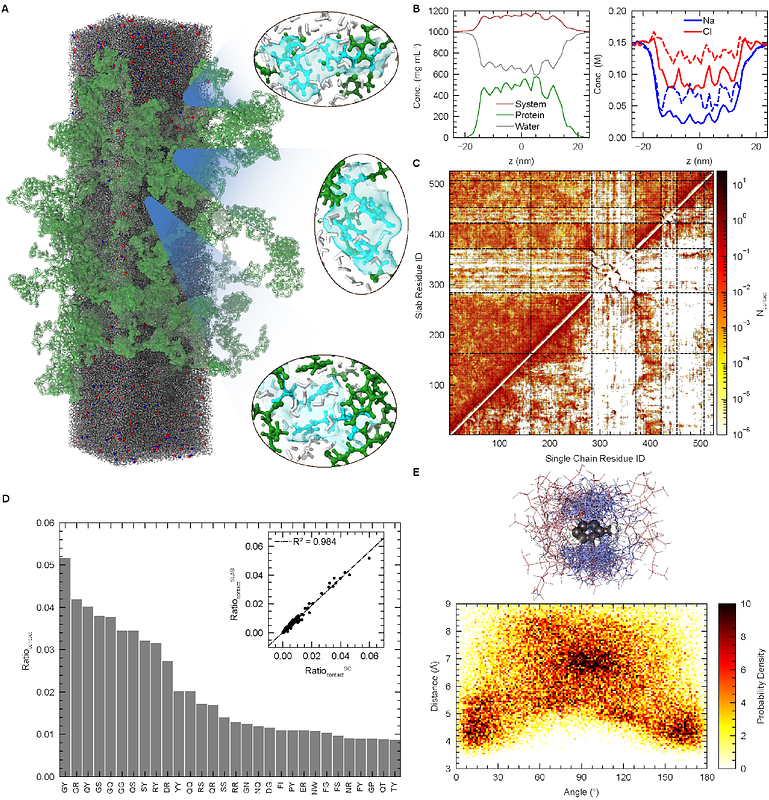
AbstractFused in Sarcoma (FUS), a multi-domain RNA-binding protein, orchestrates cellular functions through liquid-liquid phase separation (LLPS), which promotes the formation of biomolecular condensates in vivo. While crucial to understanding cellular processes, an atomic-level view of the interdomain interactions associated with full-length (FL) FUS LLPS remains challenging due to its low solubility in vitro. Here, using all-atom (AA) molecular dynamics (MD) simulations, we examined the conformational dynamics and interdomain interactions of FL FUS in both dilute and condensed phases. Comparing two modern force fields (FFs) - Amber ff03ws and ff99SBws-STQ, we found that monomer simulation ensembles generated by both FFs exhibited qualitatively similar intramolecular interaction profiles dominated by intrinsically disordered regions (IDRs). While the two folded domains minimally participated in interdomain interactions, their stabilities significantly influenced the chain dimension and led to discrepancies compared to experimental data for both FFs. We observed that the Amber ff99SBws-STQ coupled with bond parameters adopted from the Zinc Amber force field (ZAFF) maintained stable folded domains and improved estimates of the chain dimensions. Finally, a microsecond-timescale simulation of FL FUS condensate revealed an extensive network of electrostatic interactions which are strongly correlated with those that modulate the dilute phase chain dimensions. Overall, insights from our all-atom simulations illuminate the interplay between folded domain stability and IDR interactions in modulating protein conformation and phase separation, advancing our understanding of FUS-related pathologies at the molecular level and aiding in the development of new therapeutics.