Mapping Architecture of Protein complexes in Arabidopsis using XL-MS
Mapping Architecture of Protein complexes in Arabidopsis using XL-MS
Trinh, C. S.; Shrestha, R.; Conner, W. C.; Reyes, A. V.; Karunadasa, S. S.; Liu, G.; Hu, K.; Xu, S.-L.
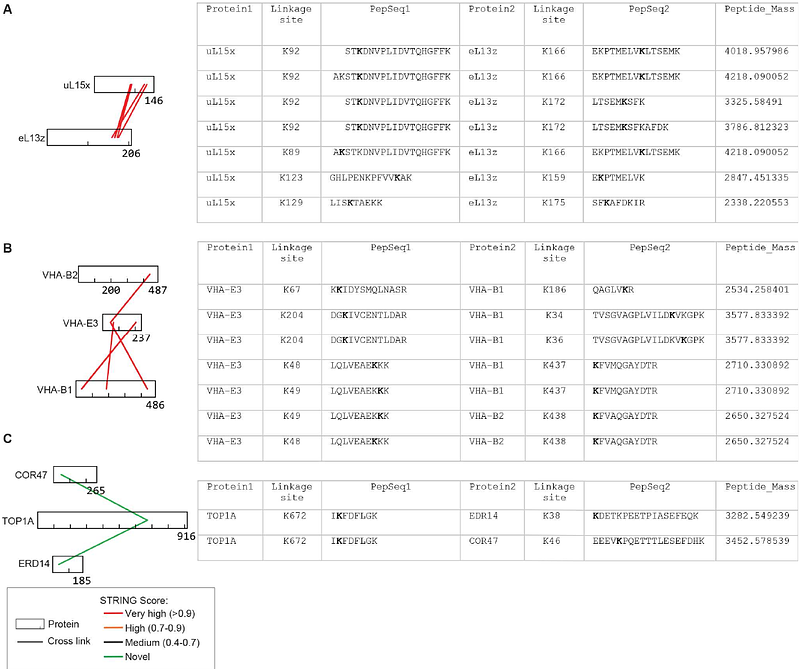
AbstractCapturing molecular machines in action is essential for understanding protein complex architecture, cellular regulation, and gene function. Here, we establish a cross-linking mass spectrometry (XL-MS) platform using the PhoX cross-linker for proteome-wide interaction mapping in Arabidopsis under semi-native conditions. From cell lysates, chloroplasts, and nuclei, we identified 47,119 unique cross-links, including 3,527 inter-protein cross-links, and constructed a high-confidence protein-protein interaction (PPI) network containing 1,229 proteins and 1,446 PPIs. In silico analysis (STRING) confirmed 637 heteromeric interactions, with the remainder representing novel interactions. Our dataset provides experimental evidence for direct interactions and defines binding interfaces. XL-MS resolved the topology of challenging proteins and revealed both conserved and unique interactions, as exemplified in photosystem complexes, ribosomes, and histone interactomes. By characterizing the histone machinery, we identified a potential novel histone modifying enzyme. This work advances protein annotation and provides new insights into cellular organization and function.