Insights into Partial Folding State of Bovine Pancreatic Trypsin Inhibitor: A Combined Molecular Dynamics Simulations, Information Theory and Molecular Graph Theory Study
Insights into Partial Folding State of Bovine Pancreatic Trypsin Inhibitor: A Combined Molecular Dynamics Simulations, Information Theory and Molecular Graph Theory Study
Kamberaj, H.
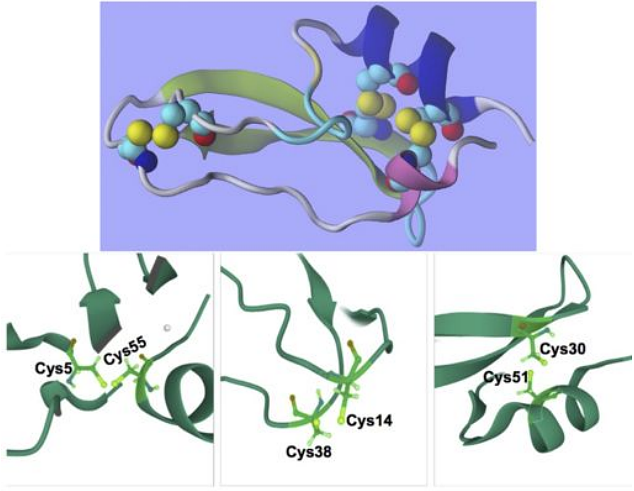
AbstractUsing a notably large amount of data in investigating physical and chemical phenomena demands new statistical and computational approaches; besides, the cross-validations require well established theoretical frameworks. This study aims to validate the statistical efficiency of alternative definitions for the information-theoretic measures, such as transfer entropy, using the (alpha; q) theoretical framework. The primary goal is to find measurements of high-order correlations that preserve information-theoretic properties of information transfer between the components of a dynamical system due to local operations. Besides, this study aims to decode the information contained in the amino acid sequence establishing a three-dimensional protein structure by comparing their physical-chemical properties with their ranked role in the protein interaction network topology using new graph-theoretic measures based on the constructed digraph models of (alpha; q) information transfer. Moreover, this study aims to use the Deep Graph Convolution Neural Networks for classifying the amino acid role in a protein structure trained upon short equilibrium structure fluctuations at sub-nanosecond time scales. In particular, this study examines the influence of disulphide bridges on the 3D protein structure of the Bovine Pancreatic Trypsin Inhibitor protein (wild type and its mutated analogue).