Comparative phenotypic, genomic and transcriptomic characterisation of two Salmonella Typhimurium strains for a first-in-human challenge model
Comparative phenotypic, genomic and transcriptomic characterisation of two Salmonella Typhimurium strains for a first-in-human challenge model
Smith, C.; Bzami, A.; Zhu, X.; White, J. A.; Roa, N.; Suvarnapunya, A. E.; Smith, E.; Rydlova, A.; Varro, R.; Killidar, Z.; Luo, L.; Perez Sepulveda, B.; Gordon, M. A.; Cooke, G. S.; Choy, R. K. M.; Hinton, J. C. D.; Gibani, M. M.
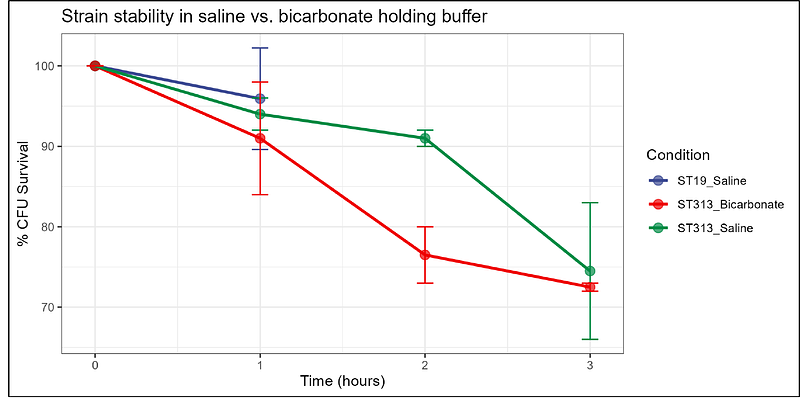
AbstractInvasive nontyphoidal Salmonella (iNTS) disease remains a major public health challenge in sub-Saharan Africa. Salmonella Typhimurium (STm) is responsible for the majority of cases, with specific lineages being associated with increased risk of bloodstream infection. We have recently developed a STm controlled human infection model (CHIM) to better understand disease pathogenesis and to provide a platform to test candidate vaccines. Selecting appropriate challenge strains is a central design consideration in developing challenge model protocols. We describe the rationale, manufacture, and detailed characterisation of two STm strains used in the first-in-human NTS CHIM. Two well-characterised STm strains were selected: 4/74 (ST19 lineage), a globally distributed diarrhoeal isolate, and D23580 (ST313 lineage 2.0), the predominant invasive pathovariant in Africa. These strains were selected based on phylogenetic relevance, antibiotic susceptibility, known provenance, and comprehensive existing virulence data. Manufacturing was conducted under Good Manufacturing Practice (GMP) conditions, with confirmation of purity, viability, and stability. Phenotypic characterisation included growth profiling, motility, colony morphology, acid and buffer sensitivity, and antibiotic susceptibility. Whole-genome sequencing confirmed genetic stability. Additionally, we investigated the effect of sodium bicarbonate exposure - used for gastric acid neutralisation - on STm virulence gene expression using transcriptomic analysis. Both strains retained expected in vitro phenotypic characteristics, including reduced motility and melibiose utilisation in D23580. Growth and viability were preserved in simulated gastrointestinal conditions and holding buffers. No unexpected mutations were detected, and both strains remained susceptible to study antibiotics. Transcriptomic profiling revealed no significant changes in key Salmonella pathogenicity island genes after sodium bicarbonate exposure, suggesting unaltered virulence at administration. These findings confirm that STm 4/74 and D23580 retain genomic and phenotypic integrity post-manufacture. The resulting challenge stocks provide the foundation for an ongoing NTS CHIM that aims to advance understanding of NTS pathogenesis and support candidate vaccine testing.