Pathogen genomic surveillance as a scalable framework for precision phage therapy
Pathogen genomic surveillance as a scalable framework for precision phage therapy
Koncz, M.; Stirling, T.; Hadj Mehdi, H.; Mehi, O.; Eszenyi, B.; Asboth, A.; Apjok, G.; Toth, A.; Orosz, L.; Vasarhelyi, B. M.; Ari, E.; Daruka, L.; Polgar, T. F.; Schneider, G.; Zalokh, S. A.; Szamel, M.; Fekete, G.; Bohar, B.; Nagy Varga, K.; Visnyovszki, A.; Szekely, E.; Licker, M. S.; Izmendi, O.; Costache, C.; Gajic, I.; Lukovic, B.; Molnar, S.; Szöcs-Gazdi, U. O.; Bozai, C.; Indreas, M.; Kristof, K.; Van der Henst, C.; Breine, A.; Pal, C.; Papp, B.; Kintses, B.
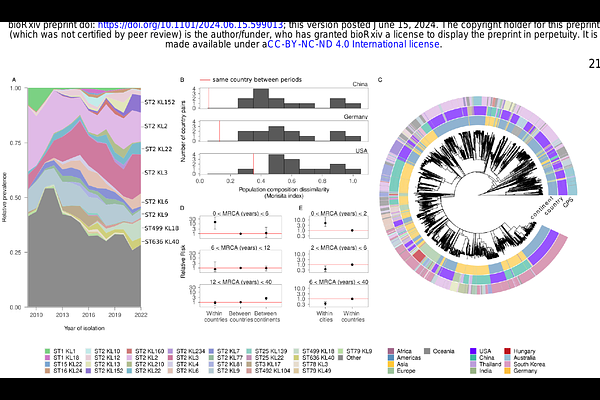
AbstractPhage therapy is gaining increasing interest in the fight against critically resistant nosocomial pathogens. However, the narrow host range of bacteriophages hampers the development of broadly effective phage therapeutics and demands precision approaches. Here we combine large-scale phylogeographical analysis with high-throughput phage typing to guide the development of precision phage cocktails targeting carbapenem-resistant Acinetobacter baumannii, a top-priority pathogen. Our analysis reveals that a few strain types dominate infections in each world region, with their geographical distribution remaining stable within six years. As we demonstrate in Eastern Europe, this spatio-temporal distribution enables preemptive preparation of region-specific phage collections that target most local infections. Finally, we showcase the efficacy of a four-phage cocktail against the most prevalent strain type in both in vitro and in vivo animal infection models. Ultimately, genomic surveillance identifies patients benefiting from the same phages across geographical scales, thus providing a scalable framework for precision phage therapy.